Escherichia coli DNA ligase B may mitigate damage from oxidative stress
- PMID: 28700629
- PMCID: PMC5507437
- DOI: 10.1371/journal.pone.0180800
Escherichia coli DNA ligase B may mitigate damage from oxidative stress
Abstract
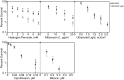
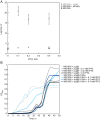
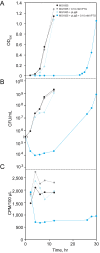
Escherichia coli encodes two DNA ligases, ligase A, which is essential under normal laboratory growth conditions, and ligase B, which is not. Here we report potential functions of ligase B. We found that across the entire Enterobacteriaceae family, ligase B is highly conserved in both amino acid identity and synteny with genes associated with oxidative stress. Deletion of ligB sensitized E. coli to specific DNA damaging agents and antibiotics resulted in a weak mutator phenotype, and decreased biofilm formation. Overexpression of ligB caused a dramatic extension of lag phase that eventually resumed normal growth. The ligase function of ligase B was not required to mediate the extended lag phase, as overexpression of a ligase-deficient ligB mutant also blocked growth. Overexpression of ligB during logarithmic growth caused an immediate block of cell growth and DNA replication, and death of about half of cells. These data support a potential role for ligase B in the base excision repair pathway or the mismatch repair pathway.
Conflict of interest statement
Figures



Similar articles
-
A second NAD(+)-dependent DNA ligase (LigB) in Escherichia coli.Nucleic Acids Res. 2001 Dec 15;29(24):4930-4. doi: 10.1093/nar/29.24.4930. Nucleic Acids Res. 2001. PMID: 11812821 Free PMC article.
-
Pol I DNA polymerases stimulate DNA end-joining by Escherichia coli DNA ligase.Biochem Biophys Res Commun. 2018 Feb 26;497(1):13-18. doi: 10.1016/j.bbrc.2018.01.165. Epub 2018 Jan 31. Biochem Biophys Res Commun. 2018. PMID: 29409896
-
Mutational analysis of Escherichia coli DNA ligase identifies amino acids required for nick-ligation in vitro and for in vivo complementation of the growth of yeast cells deleted for CDC9 and LIG4.Nucleic Acids Res. 1999 Oct 15;27(20):3953-63. doi: 10.1093/nar/27.20.3953. Nucleic Acids Res. 1999. PMID: 10497258 Free PMC article.
-
Enzymology of mitochondrial base excision repair.Prog Nucleic Acid Res Mol Biol. 2001;68:257-71. doi: 10.1016/s0079-6603(01)68105-4. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554302 Review.
-
Molecular mechanisms of DNA damage and repair: progress in plants.Crit Rev Biochem Mol Biol. 2001;36(4):337-97. doi: 10.1080/20014091074219. Crit Rev Biochem Mol Biol. 2001. PMID: 11563486 Review.
Cited by
-
Coping with Reactive Oxygen Species to Ensure Genome Stability in Escherichia coli.Genes (Basel). 2018 Nov 21;9(11):565. doi: 10.3390/genes9110565. Genes (Basel). 2018. PMID: 30469410 Free PMC article. Review.
-
Comparative genomics hints at dispensability of multiple essential genes in two Escherichia coli L-form strains.Biosci Rep. 2023 Oct 31;43(10):BSR20231227. doi: 10.1042/BSR20231227. Biosci Rep. 2023. PMID: 37819245 Free PMC article.
References
-
- Friedberg EC. DNA damage and repair. Nature. 2003;421: 436–440. doi: 10.1038/nature01408 - DOI - PubMed
-
- Sriskanda V, Shuman S. A second NAD(+)-dependent DNA ligase (LigB) in Escherichia coli. Nucleic Acids Res. 2001;29: 4930–4934. doi: 10.1093/nar/29.24.4930 - DOI - PMC - PubMed
-
- Wilkinson A, Day J, Bowater R. Bacterial DNA ligases. Mol Microbiol. 2001;40: 1241–1248. doi: 10.1046/j.1365-2958.2001.02479.x - DOI - PubMed
-
- Brötz-Oesterhelt H, Knezevic I, Bartel S, Lampe T, Warnecke-Eberz U, Ziegelbauer K, et al. Specific and potent inhibition of NAD+-dependent DNA ligase by pyridochromanones. J Biol Chem. 2003;278: 39435–39442. doi: 10.1074/jbc.M306479200 - DOI - PubMed
-
- Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, et al. Metabolomic and transcriptomic stress response of Escherichia coli. Mol Syst Biol. 2010;6 doi: 10.1038/msb.2010.18 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

