BRG1 promotes hepatocarcinogenesis by regulating proliferation and invasiveness
- PMID: 28700662
- PMCID: PMC5507512
- DOI: 10.1371/journal.pone.0180225
BRG1 promotes hepatocarcinogenesis by regulating proliferation and invasiveness
Abstract
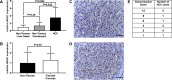
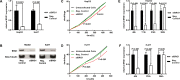
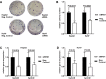
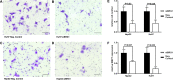
The chromatin remodeler complex SWI/SNF plays an important role in physiological and pathological processes. Brahma related gene 1(BRG1), a catalytic subunit of the SWI/SNF complex, is known to be mutated in hepatocellular carcinoma (HCC). However, its role in HCC remains unclear. Here, we investigate the role of BRG1 on cell growth and invasiveness as well as its effect on the expression of putative target genes. Expression of BRG1 was examined in human liver tissue samples and in HCC cell lines. In addition, BRG1 was silenced in human HCC cell lines to analyse cell growth and invasiveness by growth curves, colony formation assay, invasion assay and the expression of putative target genes. BRG1 was found to be significantly increased in HCC samples compared to non-HCC samples. In addition, a declined proliferation rate of BRG1-silenced human HCC cell lines was associated with a decrease of expression of cyclin family members. In line with a decreased invasiveness of BRG1-siRNA-treated human HCC cell lines, down-regulation of MMP7 was detected. These results support the hypothesis that overexpression of BRG1 increases cell growth and invasiveness in HCC. Furthermore, the data highlight cyclin B, E and MMP7 to be associated with BRG1 during hepatocarcinogenesis.
Conflict of interest statement
Figures

References
-
- Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61: 69–90. doi: 10.3322/caac.20107 - DOI - PubMed
-
- El-Serag H. B., Rudolph K. L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007; 132: 2557–2576. doi: 10.1053/j.gastro.2007.04.061 - DOI - PubMed
-
- Lee J.-S. The mutational landscape of hepatocellular carcinoma. Clin Mol Hepatol. 2015; 21: 220–229. doi: 10.3350/cmh.2015.21.3.220 - DOI - PMC - PubMed
-
- Totoki Y., Tatsuno K., Covington K.R., Ueda H., Creighton C.J., Kato M., et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014; 46: 1267–1273. doi: 10.1038/ng.3126 - DOI - PubMed
-
- Kadoch C., Hargreaves D. C., Hodges C., Elias L., Ho L., Ranish J., et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013; 45: 592–601. doi: 10.1038/ng.2628 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

