Catch Bonds at T Cell Interfaces: Impact of Surface Reorganization and Membrane Fluctuations
- PMID: 28700910
- PMCID: PMC5510709
- DOI: 10.1016/j.bpj.2017.05.023
Catch Bonds at T Cell Interfaces: Impact of Surface Reorganization and Membrane Fluctuations
Abstract
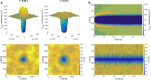
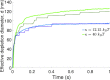
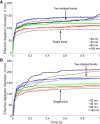
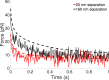
Catch bonds are characterized by average lifetimes that initially increase with increasing tensile force. Recently, they have been implicated in T cell activation, where small numbers of antigenic receptor-ligand bonds at a cell-cell interface can stimulate a T cell. Here, we use computational methods to investigate small numbers of bonds at the interface between two membranes. We characterize the time-dependent forces on the bonds in response to changes in the membrane shape and the organization of other surface molecules. We then determine the distributions of bond lifetimes using recent force-dependent lifetime data for T cell receptors bound to various ligands. Strong agonists, which exhibit catch bond behavior, are markedly more likely to remain intact than an antagonist whose average lifetime decreases with increasing force. Thermal fluctuations of the membrane shape enhance the decay of the average force on a bond, but also lead to fluctuations of the force. These fluctuations promote bond rupture, but the effect is buffered by catch bonds. When more than one bond is present, the bonds experience reduced average forces that depend on their relative positions, leading to changes in bond lifetimes. Our results highlight the importance of force-dependent binding kinetics when bonds experience time-dependent and fluctuating forces, as well as potential consequences of collective bond behavior relevant to T cell activation.
Copyright © 2017 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Dembo M., Torney D.C., Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc. R. Soc. Lond. B Biol. Sci. 1988;234:55–83. - PubMed
-
- Thomas W.E., Vogel V., Sokurenko E. Biophysics of catch bonds. Annu. Rev. Biophys. 2008;37:399–416. - PubMed
-
- Bell G.I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. - PubMed
-
- Marshall B.T., Long M., Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

