A Mutant Isoform of ObgE Causes Cell Death by Interfering with Cell Division
- PMID: 28702018
- PMCID: PMC5487468
- DOI: 10.3389/fmicb.2017.01193
A Mutant Isoform of ObgE Causes Cell Death by Interfering with Cell Division
Abstract
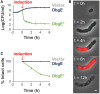
Cell division is a vital part of the cell cycle that is fundamental to all life. Despite decades of intense investigation, this process is still incompletely understood. Previously, the essential GTPase ObgE, which plays a role in a myriad of basic cellular processes (such as initiation of DNA replication, chromosome segregation, and ribosome assembly), was proposed to act as a cell cycle checkpoint in Escherichia coli by licensing chromosome segregation. We here describe the effect of a mutant isoform of ObgE (ObgE∗) that causes cell death by irreversible arrest of the cell cycle at the stage of cell division. Notably, chromosome segregation is allowed to proceed normally in the presence of ObgE∗, after which cell division is blocked. Under conditions of rapid growth, ongoing cell cycles are completed before cell cycle arrest by ObgE∗ becomes effective. However, cell division defects caused by ObgE∗ then elicit lysis through formation of membrane blebs at aberrant division sites. Based on our results, and because ObgE was previously implicated in cell cycle regulation, we hypothesize that the mutation in ObgE∗ disrupts the normal role of ObgE in cell division. We discuss how ObgE∗ could reveal more about the intricate role of wild-type ObgE in division and cell cycle control. Moreover, since Obg is widely conserved and essential for viability, also in eukaryotes, our findings might be applicable to other organisms as well.
Keywords: Obg; ObgE; cell cycle; cell cycle checkpoint; cell division; cell separation; lysis.
Figures





Similar articles
-
The bacterial cell cycle checkpoint protein Obg and its role in programmed cell death.Microb Cell. 2016 Mar 16;3(6):255-256. doi: 10.15698/mic2016.06.507. Microb Cell. 2016. PMID: 28357361 Free PMC article.
-
Deficiency of essential GTP-binding protein ObgE in Escherichia coli inhibits chromosome partition.Mol Microbiol. 2001 Sep;41(5):1037-51. doi: 10.1046/j.1365-2958.2001.02574.x. Mol Microbiol. 2001. PMID: 11555285
-
Coexpression of Escherichia coli obgE, Encoding the Evolutionarily Conserved Obg GTPase, with Ribosomal Proteins L21 and L27.J Bacteriol. 2016 Jun 13;198(13):1857-1867. doi: 10.1128/JB.00159-16. Print 2016 Jul 1. J Bacteriol. 2016. PMID: 27137500 Free PMC article.
-
The structure-function analysis of Obg-like GTPase proteins along the evolutionary tree from bacteria to humans.Genes Cells. 2022 Jul;27(7):469-481. doi: 10.1111/gtc.12942. Epub 2022 May 24. Genes Cells. 2022. PMID: 35610748 Free PMC article. Review.
-
Bacterial Obg proteins: GTPases at the nexus of protein and DNA synthesis.Crit Rev Microbiol. 2014 Aug;40(3):207-24. doi: 10.3109/1040841X.2013.776510. Epub 2013 Mar 28. Crit Rev Microbiol. 2014. PMID: 23537324 Review.
Cited by
-
Image-Based Dynamic Phenotyping Reveals Genetic Determinants of Filamentation-Mediated β-Lactam Tolerance.Front Microbiol. 2020 Mar 13;11:374. doi: 10.3389/fmicb.2020.00374. eCollection 2020. Front Microbiol. 2020. PMID: 32231648 Free PMC article.
-
The role of the essential GTPase ObgE in regulating lipopolysaccharide synthesis in Escherichia coli.Nat Commun. 2024 Nov 8;15(1):9684. doi: 10.1038/s41467-024-53980-1. Nat Commun. 2024. PMID: 39516202 Free PMC article.
-
Overproduction of a Dominant Mutant of the Conserved Era GTPase Inhibits Cell Division in Escherichia coli.J Bacteriol. 2020 Oct 8;202(21):e00342-20. doi: 10.1128/JB.00342-20. Print 2020 Oct 8. J Bacteriol. 2020. PMID: 32817092 Free PMC article.
-
High-throughput time-resolved morphology screening in bacteria reveals phenotypic responses to antibiotics.Commun Biol. 2019 Jul 23;2:269. doi: 10.1038/s42003-019-0480-9. eCollection 2019. Commun Biol. 2019. PMID: 31341968 Free PMC article.
-
In silico insight of cell-death-related proteins in photosynthetic cyanobacteria.Arch Microbiol. 2022 Jul 21;204(8):511. doi: 10.1007/s00203-022-03130-2. Arch Microbiol. 2022. PMID: 35864385 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

