Molecular indices of viral disease development in wild migrating salmon†
- PMID: 28702195
- PMCID: PMC5499884
- DOI: 10.1093/conphys/cox036
Molecular indices of viral disease development in wild migrating salmon†
Abstract
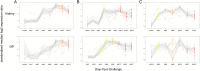
Infectious diseases can impact the physiological performance of individuals, including their mobility, visual acuity, behavior and tolerance and ability to effectively respond to additional stressors. These physiological effects can influence competitiveness, social hierarchy, habitat usage, migratory behavior and risk to predation, and in some circumstances, viability of populations. While there are multiple means of detecting infectious agents (microscopy, culture, molecular assays), the detection of infectious diseases in wild populations in circumstances where mortality is not observable can be difficult. Moreover, if infection-related physiological compromise leaves individuals vulnerable to predation, it may be rare to observe wildlife in a late stage of disease. Diagnostic technologies designed to diagnose cause of death are not always sensitive enough to detect early stages of disease development in live-sampled organisms. Sensitive technologies that can differentiate agent carrier states from active disease states are required to demonstrate impacts of infectious diseases in wild populations. We present the discovery and validation of salmon host transcriptional biomarkers capable of distinguishing fish in an active viral disease state [viral disease development (VDD)] from those carrying a latent viral infection, and viral versus bacterial disease states. Biomarker discovery was conducted through meta-analysis of published and in-house microarray data, and validation performed on independent datasets including disease challenge studies and farmed salmon diagnosed with various viral, bacterial and parasitic diseases. We demonstrate that the VDD biomarker panel is predictive of disease development across RNA-viral species, salmon species and salmon tissues, and can recognize a viral disease state in wild-migrating salmon. Moreover, we show that there is considerable overlap in the biomarkers resolved in our study in salmon with those based on similar human viral influenza research, suggesting a highly conserved suite of host genes associated with viral disease that may be applicable across a broad range of vertebrate taxa.
Keywords: aquaculture; disease biomarkers; host transcriptome; salmon; viral disease; wild populations.
Figures











Similar articles
-
De novo assembly of Sockeye salmon kidney transcriptomes reveal a limited early response to piscine reovirus with or without infectious hematopoietic necrosis virus superinfection.BMC Genomics. 2016 Nov 2;17(1):848. doi: 10.1186/s12864-016-3196-y. BMC Genomics. 2016. PMID: 27806699 Free PMC article.
-
Infectious hematopoietic necrosis virus (IHNV) persistence in Sockeye Salmon: influence on brain transcriptome and subsequent response to the viral mimic poly(I:C).BMC Genomics. 2015 Aug 26;16(1):634. doi: 10.1186/s12864-015-1759-y. BMC Genomics. 2015. PMID: 26306576 Free PMC article.
-
Isolation and identification of infectious salmon anaemia virus (ISAV) from Coho salmon in Chile.Dis Aquat Organ. 2001 May 4;45(1):9-18. doi: 10.3354/dao045009. Dis Aquat Organ. 2001. PMID: 11411649
-
An evaluation of the effects of conservation and fishery enhancement hatcheries on wild populations of salmon.Adv Mar Biol. 2007;53:61-194. doi: 10.1016/S0065-2881(07)53002-6. Adv Mar Biol. 2007. PMID: 17936136 Review.
-
Infectious diseases affect marine fisheries and aquaculture economics.Ann Rev Mar Sci. 2015;7:471-96. doi: 10.1146/annurev-marine-010814-015646. Epub 2014 Sep 12. Ann Rev Mar Sci. 2015. PMID: 25251276 Review.
Cited by
-
Using transcriptomics to predict and visualize disease status in bighorn sheep (Ovis canadensis).Conserv Physiol. 2022 Jul 3;10(1):coac046. doi: 10.1093/conphys/coac046. eCollection 2022. Conserv Physiol. 2022. PMID: 35795016 Free PMC article.
-
Baseline Gene Expression Levels in Falkland-Malvinas Island Penguins: Towards a New Monitoring Paradigm.Life (Basel). 2022 Feb 9;12(2):258. doi: 10.3390/life12020258. Life (Basel). 2022. PMID: 35207543 Free PMC article.
-
Infectious agents and their physiological correlates in early marine Chinook salmon (Oncorhynchus tshawytscha).Conserv Physiol. 2023 May 19;11(1):coad031. doi: 10.1093/conphys/coad031. eCollection 2023. Conserv Physiol. 2023. PMID: 37701371 Free PMC article.
-
Evidence of a hydraulically challenging reach serving as a barrier for the upstream migration of infection-burdened adult steelhead.Conserv Physiol. 2019 Jun 6;7(1):coz023. doi: 10.1093/conphys/coz023. eCollection 2019. Conserv Physiol. 2019. PMID: 31191906 Free PMC article.
-
Genomes reveal genetic diversity of Piscine orthoreovirus in farmed and free-ranging salmonids from Canada and USA.Virus Evol. 2020 Jul 31;6(2):veaa054. doi: 10.1093/ve/veaa054. eCollection 2020 Jul. Virus Evol. 2020. PMID: 33381304 Free PMC article.
References
-
- Anderson RM, May RM (1986) The invasion, persistence and spread of infectious diseases within animal and plant communities. Philos Trans R Ser B 314: 533–570. - PubMed
-
- Bakke T, Harris P (1998) Diseases and parasites in wild Atlantic salmon (Salmo salar) populations. Can J Fish Aquat Sci 55: 247–266.
-
- Beaulaurier J, Bickford N, Gregg JL, Grady CA, Gannam AL, Winton JR, Hershberger PK (2012) Susceptibility of Pacific herring to viral hemorrhagic septicemia is influenced by diet. J Aquat Anim Health 24: 43–48. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

