In Vivo Delivery and Activation of Masked Fluorogenic Hydrolase Substrates by Endogenous Hydrolases in C. elegans
- PMID: 28703362
- PMCID: PMC5605452
- DOI: 10.1002/cbic.201700278
In Vivo Delivery and Activation of Masked Fluorogenic Hydrolase Substrates by Endogenous Hydrolases in C. elegans
Abstract
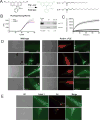
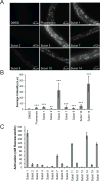
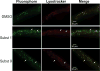
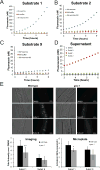
Protein expression and localization are often studied in vivo by tagging molecules with green fluorescent protein (GFP), yet subtle changes in protein levels are not easily detected. To develop a sensitive in vivo method to amplify fluorescence signals and allow cell-specific quantification of protein abundance changes, we sought to apply an enzyme-activated cellular fluorescence system in vivo by delivering ester-masked fluorophores to Caenorhabditis elegans neurons expressing porcine liver esterase (PLE). To aid uptake into sensory neuron membranes, we synthesized two novel fluorogenic hydrolase substrates with long hydrocarbon tails. Recombinant PLE activated these fluorophores in vitro. In vivo activation occurred in sensory neurons, along with potent activation in intestinal lysosomes quantifiable by imaging and microplate and partially attributable to gut esterase 1 (GES-1) activity. These data demonstrate the promise of biorthogonal hydrolases and their fluorogenic substrates as in vivo neuronal imaging tools and for characterizing endogenous C. elegans hydrolase substrate specificities.
Keywords: C. elegans; bioorthogonal; fluorescent probes; hydrolases; imaging agents.
© 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

