The emerging role of alternative splicing in senescence and aging
- PMID: 28703423
- PMCID: PMC5595669
- DOI: 10.1111/acel.12646
The emerging role of alternative splicing in senescence and aging
Abstract
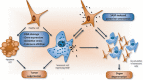
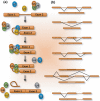
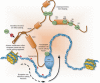
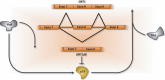
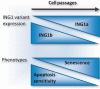
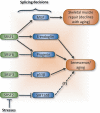
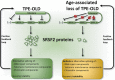
Deregulation of precursor mRNA splicing is associated with many illnesses and has been linked to age-related chronic diseases. Here we review recent progress documenting how defects in the machinery that performs intron removal and controls splice site selection contribute to cellular senescence and organismal aging. We discuss the functional association linking p53, IGF-1, SIRT1, and ING-1 splice variants with senescence and aging, and review a selection of splicing defects occurring in accelerated aging (progeria), vascular aging, and Alzheimer's disease. Overall, it is becoming increasingly clear that changes in the activity of splicing factors and in the production of key splice variants can impact cellular senescence and the aging phenotype.
Keywords: RNA; RNA binding proteins; aging; alternative splicing; pre-mRNA; senescence; splice variants; splicing.
© 2017 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures

References
-
- Abad M, Moreno A, Palacios A, Narita M, Blanco F, Moreno‐Bueno G, Narita M, Palmero I (2011) The tumor suppressor ING1 contributes to epigenetic control of cellular senescence. Aging Cell 10, 158–171. - PubMed
-
- Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, Athineos D, Kang T‐W, Lasitschka F, Andrulis M, Pascual G, Morris KJ, Khan S, Jin H, Dharmalingam G, Snijders AP, Carroll T, Capper D, Pritchard C, Inman GJ, Longerich T, Sansom OJ, Benitah SA, Zender L, Gil J (2013) A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 15, 978–990. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

