Do social insects support Haig's kin theory for the evolution of genomic imprinting?
- PMID: 28703654
- PMCID: PMC5739101
- DOI: 10.1080/15592294.2017.1348445
Do social insects support Haig's kin theory for the evolution of genomic imprinting?
Abstract
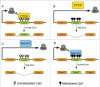
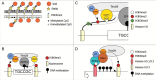
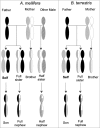
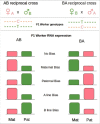
Although numerous imprinted genes have been described in several lineages, the phenomenon of genomic imprinting presents a peculiar evolutionary problem. Several hypotheses have been proposed to explain gene imprinting, the most supported being Haig's kinship theory. This theory explains the observed pattern of imprinting and the resulting phenotypes as a competition for resources between related individuals, but despite its relevance it has not been independently tested. Haig's theory predicts that gene imprinting should be present in eusocial insects in many social scenarios. These lineages are therefore ideal for testing both the theory's predictions and the mechanism of gene imprinting. Here we review the behavioral evidence of genomic imprinting in eusocial insects, the evidence of a mechanism for genomic imprinting and finally we evaluate recent results showing parent of origin allele specific expression in honeybees in the light of Haig's theory.
Keywords: Epigenetics; Haig's theory; evolution; genomic imprinting; social insects.
Figures
References
-
- Haig D. The kinship theory of genomic imprinting. Annual review of ecology and systematics 2000; 9-32 doi:10.1146/annurev.ecolsys.31.1.9 - DOI
-
- Hurst LD. Evolutionary theories of genomic imprinting In Reik W. and Surani A., editors, Genomic imprinting. IRL Press, Oxford, 1997.
-
- Patten MM, Ross L, Curley JP, Queller DC, Bonduriansky R, Wolf JB. The evolution of genomic imprinting: theories, predictions and empirical tests. Heredity 2014; 113(2):119-28; PMID:24755983; https://doi.org/10.1038/hdy.2014.29 - DOI - PMC - PubMed
-
- Queller DC. Theory of genomic imprinting conflict in social insects. BMC Evol Biol 2003; 3(1):1; PMID:12515583; https://doi.org/10.1186/1471-2148-3-15 - DOI - PMC - PubMed
-
- Spencer HG, Clark AG. Non-conflict theories for the evolution of genomic imprinting. Heredity 2014; 113(2):112-8; PMID:24398886; https://doi.org/10.1038/hdy.2013.129 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
