Development of Phosphorothioate DNA and DNA Thioaptamers
- PMID: 28703779
- PMCID: PMC5618299
- DOI: 10.3390/biomedicines5030041
Development of Phosphorothioate DNA and DNA Thioaptamers
Abstract
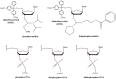
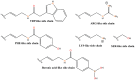
Nucleic acid aptamers are short RNA- or DNA-based affinity reagents typically selected from combinatorial libraries to bind to a specific target such as a protein, a small molecule, whole cells or even animals. Aptamers have utility in the development of diagnostic, imaging and therapeutic applications due to their size, physico-chemical nature and ease of synthesis and modification to suit the application. A variety of oligonucleotide modifications have been used to enhance the stability of aptamers from nuclease degradation in vivo. The non-bridging oxygen atoms of the phosphodiester backbones of RNA and DNA aptamers can be substituted with one or two sulfur atoms, resulting in thioaptamers with phosphorothioate or phosphorodithioate linkages, respectively. Such thioaptamers are known to have increased binding affinity towards their target, as well as enhanced resistance to nuclease degradation. In this review, we discuss the development of phosphorothioate chemistry and thioaptamers, with a brief review of selection methods.
Keywords: DNA aptamer; X-aptamer; thioaptamer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

