Inhibition of autophagy as a treatment strategy for p53 wild-type acute myeloid leukemia
- PMID: 28703806
- PMCID: PMC5550863
- DOI: 10.1038/cddis.2017.317
Inhibition of autophagy as a treatment strategy for p53 wild-type acute myeloid leukemia
Abstract
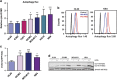
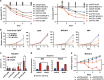
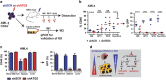
Here we have explored whether inhibition of autophagy can be used as a treatment strategy for acute myeloid leukemia (AML). Steady-state autophagy was measured in leukemic cell lines and primary human CD34+ AML cells with a large variability in basal autophagy between AMLs observed. The autophagy flux was higher in AMLs classified as poor risk, which are frequently associated with TP53 mutations (TP53mut), compared with favorable- and intermediate-risk AMLs. In addition, the higher flux was associated with a higher expression level of several autophagy genes, but was not affected by alterations in p53 expression by knocking down p53 or overexpression of wild-type p53 or p53R273H. AML CD34+ cells were more sensitive to the autophagy inhibitor hydroxychloroquine (HCQ) than normal bone marrow CD34+ cells. Similar, inhibition of autophagy by knockdown of ATG5 or ATG7 triggered apoptosis, which coincided with increased expression of p53. In contrast to wild-type p53 AML (TP53wt), HCQ treatment did not trigger a BAX and PUMA-dependent apoptotic response in AMLs harboring TP53mut. To further characterize autophagy in the leukemic stem cell-enriched cell fraction AML CD34+ cells were separated into ROSlow and ROShigh subfractions. The immature AML CD34+-enriched ROSlow cells maintained higher basal autophagy and showed reduced survival upon HCQ treatment compared with ROShigh cells. Finally, knockdown of ATG5 inhibits in vivo maintenance of AML CD34+ cells in NSG mice. These results indicate that targeting autophagy might provide new therapeutic options for treatment of AML since it affects the immature AML subfraction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Shen Y, Zhu Y-M, Fan X, Shi JY, Wang QR, Yan XJ et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood 2011; 118: 5593–5603. - PubMed
-
- Wouters BJ, Delwel R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood 2016; 127: 42–52. - PubMed
-
- Rose D, Haferlach T, Schnittger S, Takahashi N, Yamashita T. Subtype-specific patterns of molecular mutations in acute myeloid leukemia. Leukemia 2017; 31: 11–17. - PubMed
-
- Testa U, Riccioni R. Deregulation of apoptosis in acute myeloid leukemia. Haematologica 2007; 92: 81–94. - PubMed
-
- Bosman MCJ, Schepers H, Jaques J, Brouwers-Vos AZ, Quax WJ, Schuringa JJ et al. The TAK1-NF-κB axis as therapeutic target for AML. Blood 2014; 124: 3130–3140. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

