Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides
- PMID: 28704043
- PMCID: PMC6443407
- DOI: 10.1021/jacs.7b01283
Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides
Abstract
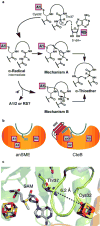
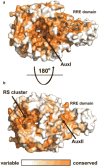
Sactipeptides are ribosomally synthesized peptides that contain a characteristic thioether bridge (sactionine bond) that is installed posttranslationally and is absolutely required for their antibiotic activity. Sactipeptide biosynthesis requires a unique family of radical SAM enzymes, which contain multiple [4Fe-4S] clusters, to form the requisite thioether bridge between a cysteine and the α-carbon of an opposing amino acid through radical-based chemistry. Here we present the structure of the sactionine bond-forming enzyme CteB, from Clostridium thermocellum ATCC 27405, with both SAM and an N-terminal fragment of its peptidyl-substrate at 2.04 Å resolution. CteB has the (β/α)6-TIM barrel fold that is characteristic of radical SAM enzymes, as well as a C-terminal SPASM domain that contains two auxiliary [4Fe-4S] clusters. Importantly, one [4Fe-4S] cluster in the SPASM domain exhibits an open coordination site in absence of peptide substrate, which is coordinated by a peptidyl-cysteine residue in the bound state. The crystal structure of CteB also reveals an accessory N-terminal domain that has high structural similarity to a recently discovered motif present in several enzymes that act on ribosomally synthesized and post-translationally modified peptides (RiPPs), known as a RiPP precursor peptide recognition element (RRE). This crystal structure is the first of a sactionine bond forming enzyme and sheds light on structures and mechanisms of other members of this class such as AlbA or ThnB.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




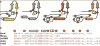
References
-
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Nat Prod Rep. 2012;30(1):108. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

