Mouse models of 17q21.31 microdeletion and microduplication syndromes highlight the importance of Kansl1 for cognition
- PMID: 28704368
- PMCID: PMC5531616
- DOI: 10.1371/journal.pgen.1006886
Mouse models of 17q21.31 microdeletion and microduplication syndromes highlight the importance of Kansl1 for cognition
Abstract
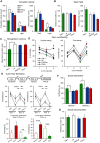
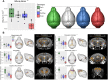
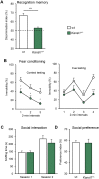
Koolen-de Vries syndrome (KdVS) is a multi-system disorder characterized by intellectual disability, friendly behavior, and congenital malformations. The syndrome is caused either by microdeletions in the 17q21.31 chromosomal region or by variants in the KANSL1 gene. The reciprocal 17q21.31 microduplication syndrome is associated with psychomotor delay, and reduced social interaction. To investigate the pathophysiology of 17q21.31 microdeletion and microduplication syndromes, we generated three mouse models: 1) the deletion (Del/+); or 2) the reciprocal duplication (Dup/+) of the 17q21.31 syntenic region; and 3) a heterozygous Kansl1 (Kans1+/-) model. We found altered weight, general activity, social behaviors, object recognition, and fear conditioning memory associated with craniofacial and brain structural changes observed in both Del/+ and Dup/+ animals. By investigating hippocampus function, we showed synaptic transmission defects in Del/+ and Dup/+ mice. Mutant mice with a heterozygous loss-of-function mutation in Kansl1 displayed similar behavioral and anatomical phenotypes compared to Del/+ mice with the exception of sociability phenotypes. Genes controlling chromatin organization, synaptic transmission and neurogenesis were upregulated in the hippocampus of Del/+ and Kansl1+/- animals. Our results demonstrate the implication of KANSL1 in the manifestation of KdVS phenotypes and extend substantially our knowledge about biological processes affected by these mutations. Clear differences in social behavior and gene expression profiles between Del/+ and Kansl1+/- mice suggested potential roles of other genes affected by the 17q21.31 deletion. Together, these novel mouse models provide new genetic tools valuable for the development of therapeutic approaches.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Vulto-van Silfhout AT, Hehir-Kwa JY, van Bon BW, Schuurs-Hoeijmakers JH, Meader S, Hellebrekers CJ, et al. Clinical significance of de novo and inherited copy-number variation. Hum Mutat. 2013;34(12):1679–87. Epub 2013/09/17. doi: 10.1002/humu.22442 . - DOI - PubMed
-
- Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, Bosco P, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nature genetics. 2014;46(10):1063–71. Epub 2014/09/15. doi: 10.1038/ng.3092 ; - DOI - PMC - PubMed
-
- Koolen DA, Pfundt R, Linda K, Beunders G, Veenstra-Knol HE, Conta JH, et al. The Koolen-de Vries syndrome: a phenotypic comparison of patients with a 17q21.31 microdeletion versus a KANSL1 sequence variant. Eur J Hum Genet. 2015. doi: 10.1038/ejhg.2015.178 . - DOI - PMC - PubMed
-
- Koolen DA, Vissers LE, Pfundt R, de Leeuw N, Knight SJ, Regan R, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nature genetics. 2006;38(9):999–1001. Epub 2006/08/15. doi: 10.1038/ng1853 . - DOI - PubMed
-
- Benda CE, Mann GV. The serum cholesterol and lipoprotein levels in mongolism. J Pediatr. 1955;46(1):49–53. . - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

