Rapid identification of genes controlling virulence and immunity in malaria parasites
- PMID: 28704525
- PMCID: PMC5507557
- DOI: 10.1371/journal.ppat.1006447
Rapid identification of genes controlling virulence and immunity in malaria parasites
Abstract
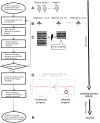
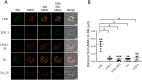
Identifying the genetic determinants of phenotypes that impact disease severity is of fundamental importance for the design of new interventions against malaria. Here we present a rapid genome-wide approach capable of identifying multiple genetic drivers of medically relevant phenotypes within malaria parasites via a single experiment at single gene or allele resolution. In a proof of principle study, we found that a previously undescribed single nucleotide polymorphism in the binding domain of the erythrocyte binding like protein (EBL) conferred a dramatic change in red blood cell invasion in mutant rodent malaria parasites Plasmodium yoelii. In the same experiment, we implicated merozoite surface protein 1 (MSP1) and other polymorphic proteins, as the major targets of strain-specific immunity. Using allelic replacement, we provide functional validation of the substitution in the EBL gene controlling the growth rate in the blood stages of the parasites.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Nair S, Williams JT, Brockman A, Paiphun L, Mayxay M, Newton PN, Guthmann JP, Smithuis FM, Hien TT, White NJ, Nosten F, Anderson TJ. A selective sweep driven by pyrimethamine treatment in southeast asian malaria parasites. Mol Biol Evol. 2003. September;20(9):1526–1536. 10.1093/molbev/msg162 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

