Receptor activity-modifying protein dependent and independent activation mechanisms in the coupling of calcitonin gene-related peptide and adrenomedullin receptors to Gs
- PMID: 28705698
- PMCID: PMC5609567
- DOI: 10.1016/j.bcp.2017.07.005
Receptor activity-modifying protein dependent and independent activation mechanisms in the coupling of calcitonin gene-related peptide and adrenomedullin receptors to Gs
Abstract
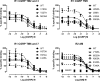
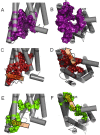
Calcitonin gene-related peptide (CGRP) or adrenomedullin (AM) receptors are heteromers of the calcitonin receptor-like receptor (CLR), a class B G protein-coupled receptor, and one of three receptor activity-modifying proteins (RAMPs). How CGRP and AM activate CLR and how this process is modulated by RAMPs is unclear. We have defined how CGRP and AM induce Gs-coupling in CLR-RAMP heteromers by measuring the effect of targeted mutagenesis in the CLR transmembrane domain on cAMP production, modeling the active state conformations of CGRP and AM receptors in complex with the Gs C-terminus and conducting molecular dynamics simulations in an explicitly hydrated lipidic bilayer. The largest effects on receptor signaling were seen with H295A5.40b, I298A5.43b, L302A5.47b, N305A5.50b, L345A6.49b and E348A6.52b, F349A6.53b and H374A7.47b (class B numbering in superscript). Many of these residues are likely to form part of a group in close proximity to the peptide binding site and link to a network of hydrophilic and hydrophobic residues, which undergo rearrangements to facilitate Gs binding. Residues closer to the extracellular loops displayed more pronounced RAMP or ligand-dependent effects. Mutation of H3747.47b to alanine increased AM potency 100-fold in the CGRP receptor. The molecular dynamics simulation showed that TM5 and TM6 pivoted around TM3. The data suggest that hydrophobic interactions are more important for CLR activation than other class B GPCRs, providing new insights into the mechanisms of activation of this class of receptor. Furthermore the data may aid in the understanding of how RAMPs modulate the signaling of other class B GPCRs.
Keywords: Adrenomedullin; Adrenomedullin (PubChem CID: 56841671); Calcitonin gene-related peptide; Calcitonin gene-related peptide (PubChem CID: 56841902); GPCR; Molecular dynamics; Molecular modeling; Receptor activity-modifying protein; cAMP (PubChem CID: 6076).
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

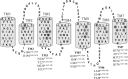





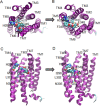


References
-
- Archbold J.K., Flanagan J.U., Watkins H.A., Gingell J.J., Hay D.L. Structural insights into RAMP modification of secretin family G protein-coupled receptors: implications for drug development. Trends Pharmacol. Sci. 2011;32(10):591–600. - PubMed
-
- Bailey R.J., Hay D.L. Pharmacology of the human CGRP1 receptor in Cos 7 cells. Peptides. 2006;27(6):1367–1375. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

