Cellular heterogeneity in the ureteric progenitor niche and distinct profiles of branching morphogenesis in organ development
- PMID: 28705898
- PMCID: PMC5611955
- DOI: 10.1242/dev.149112
Cellular heterogeneity in the ureteric progenitor niche and distinct profiles of branching morphogenesis in organ development
Abstract
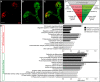
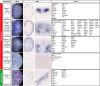
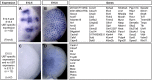
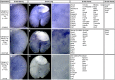
Branching morphogenesis creates arborized epithelial networks. In the mammalian kidney, an epithelial progenitor pool at ureteric branch tips (UBTs) creates the urine-transporting collecting system. Using region-specific mouse reporter strains, we performed an RNA-seq screen, identifying tip- and stalk-enriched gene sets in the developing collecting duct system. Detailed in situ hybridization studies of tip-enriched predictions identified UBT-enriched gene sets conserved between the mouse and human kidney. Comparative spatial analysis of their UBT niche expression highlighted distinct patterns of gene expression revealing novel molecular heterogeneity within the UBT progenitor population. To identify kidney-specific and shared programs of branching morphogenesis, comparative expression studies on the developing mouse lung were combined with in silico analysis of the developing mouse salivary gland. These studies highlight a shared gene set with multi-organ tip enrichment and a gene set specific to UBTs. This comprehensive analysis extends our current understanding of the ureteric branch tip niche.
Keywords: Branching morphogenesis; RNA-seq; Tip progenitor; Ureteric bud.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures



References
-
- Blomqvist S. R., Vidarsson H., Fitzgerald S., Johansson B. R., Ollerstam A., Brown R., Persson A. E. G., Bergström G. G. and Enerbäck S. (2004). Distal renal tubular acidosis in mice that lack the forkhead transcription factor Foxi1. J. Clin. Invest. 113, 1560-1570. 10.1172/JCI20665 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

