TGF-β inhibits alveolar protein transport by promoting shedding, regulated intramembrane proteolysis, and transcriptional downregulation of megalin
- PMID: 28705909
- PMCID: PMC6148198
- DOI: 10.1152/ajplung.00569.2016
TGF-β inhibits alveolar protein transport by promoting shedding, regulated intramembrane proteolysis, and transcriptional downregulation of megalin
Abstract
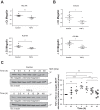
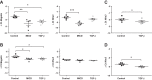
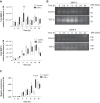
Disruption of the alveolar-capillary barrier is a hallmark of acute respiratory distress syndrome (ARDS) that leads to the accumulation of protein-rich edema in the alveolar space, often resulting in comparable protein concentrations in alveolar edema and plasma and causing deleterious remodeling. Patients who survive ARDS have approximately three times lower protein concentrations in the alveolar edema than nonsurvivors; thus the ability to remove excess protein from the alveolar space may be critical for a positive outcome. We have recently shown that clearance of albumin from the alveolar space is mediated by megalin, a 600-kDa transmembrane endocytic receptor and member of the low-density lipoprotein receptor superfamily. In the currents study, we investigate the molecular mechanisms by which transforming growth factor-β (TGF-β), a key molecule of ARDS pathogenesis, drives downregulation of megalin expression and function. TGF-β treatment led to shedding and regulated intramembrane proteolysis of megalin at the cell surface and to a subsequent increase in intracellular megalin COOH-terminal fragment abundance resulting in transcriptional downregulation of megalin. Activity of classical protein kinase C enzymes and γ-secretase was required for the TGF-β-induced megalin downregulation. Furthermore, TGF-β-induced shedding of megalin was mediated by matrix metalloproteinases (MMPs)-2, -9, and -14. Silencing of either of these MMPs stabilized megalin at the cell surface after TGF-β treatment and restored normal albumin transport. Moreover, a direct interaction of megalin with MMP-2 and -14 was demonstrated, suggesting that these MMPs may function as novel sheddases of megalin. Further understanding of these mechanisms may lead to novel therapeutic approaches for the treatment of ARDS.
Keywords: alveolar protein transport; matrix metalloproteinases; megalin; receptor shedding; regulated intramembrane proteolysis.
Copyright © 2017 the American Physiological Society.
Figures








Similar articles
-
Influenza virus decreases albumin uptake and megalin expression in alveolar epithelial cells.Front Immunol. 2023 Sep 1;14:1260973. doi: 10.3389/fimmu.2023.1260973. eCollection 2023. Front Immunol. 2023. PMID: 37727782 Free PMC article.
-
Linking receptor-mediated endocytosis and cell signaling: evidence for regulated intramembrane proteolysis of megalin in proximal tubule.J Biol Chem. 2004 Aug 13;279(33):34302-10. doi: 10.1074/jbc.M405608200. Epub 2004 Jun 4. J Biol Chem. 2004. PMID: 15180987
-
Restoration of Megalin-Mediated Clearance of Alveolar Protein as a Novel Therapeutic Approach for Acute Lung Injury.Am J Respir Cell Mol Biol. 2017 Nov;57(5):589-602. doi: 10.1165/rcmb.2016-0358OC. Am J Respir Cell Mol Biol. 2017. PMID: 28678521 Free PMC article.
-
Regulated intramembrane proteolysis of megalin: linking urinary protein and gene regulation in proximal tubule?Kidney Int. 2006 May;69(10):1717-21. doi: 10.1038/sj.ki.5000298. Kidney Int. 2006. PMID: 16557231 Review.
-
New insights into the roles of megalin/LRP2 and the regulation of its functional expression.Biol Res. 2011;44(1):89-105. doi: 10.4067/S0716-97602011000100012. Epub 2011 May 11. Biol Res. 2011. PMID: 21720686 Review.
Cited by
-
Participation of OCRL1, and APPL1, in the expression, proteolysis, phosphorylation and endosomal trafficking of megalin: Implications for Lowe Syndrome.Front Cell Dev Biol. 2022 Oct 20;10:911664. doi: 10.3389/fcell.2022.911664. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36340038 Free PMC article.
-
Alveolar-capillary endocytosis and trafficking in acute lung injury and acute respiratory distress syndrome.Front Immunol. 2024 Mar 12;15:1360370. doi: 10.3389/fimmu.2024.1360370. eCollection 2024. Front Immunol. 2024. PMID: 38533500 Free PMC article. Review.
-
Potential therapeutic effects of interleukin-35 on the differentiation of naïve T cells into Helios+Foxp3+ Tregs in clinical and experimental acute respiratory distress syndrome.Mol Immunol. 2021 Apr;132:236-249. doi: 10.1016/j.molimm.2021.01.009. Epub 2021 Jan 16. Mol Immunol. 2021. PMID: 33494935 Free PMC article.
-
Influenza virus decreases albumin uptake and megalin expression in alveolar epithelial cells.Front Immunol. 2023 Sep 1;14:1260973. doi: 10.3389/fimmu.2023.1260973. eCollection 2023. Front Immunol. 2023. PMID: 37727782 Free PMC article.
-
Clathrin-Mediated Albumin Clearance in Alveolar Epithelial Cells of Murine Precision-Cut Lung Slices.Int J Mol Sci. 2023 Jan 31;24(3):2644. doi: 10.3390/ijms24032644. Int J Mol Sci. 2023. PMID: 36768968 Free PMC article.
References
-
- Bachofen M, Weibel ER. Structural alterations of lung parenchyma in the adult respiratory distress syndrome. Clin Chest Med : 35–56, 1982. - PubMed
-
- Berthiaume Y, Albertine KH, Grady M, Fick G, Matthay MA. Protein clearance from the air spaces and lungs of unanesthetized sheep over 144 h. J Appl Physiol (1985) : 1887–1897, 1989. - PubMed
-
- Biemesderfer D, Pizzonia J, Abu-Alfa A, Exner M, Reilly R, Igarashi P, Aronson PS. NHE3: a Na+/H+ exchanger isoform of renal brush border. Am J Physiol : F736–F742, 1993. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

