Microarray analysis of bone marrow lesions in osteoarthritis demonstrates upregulation of genes implicated in osteochondral turnover, neurogenesis and inflammation
- PMID: 28705915
- PMCID: PMC5629942
- DOI: 10.1136/annrheumdis-2017-211396
Microarray analysis of bone marrow lesions in osteoarthritis demonstrates upregulation of genes implicated in osteochondral turnover, neurogenesis and inflammation
Abstract
Objective: Bone marrow lesions (BMLs) are well described in osteoarthritis (OA) using MRI and are associated with pain, but little is known about their pathological characteristics and gene expression. We evaluated BMLs using novel tissue analysis tools to gain a deeper understanding of their cellular and molecular expression.
Methods: We recruited 98 participants, 72 with advanced OA requiring total knee replacement (TKR), 12 with mild OA and 14 non-OA controls. Participants were assessed for pain (using Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC)) and with a knee MRI (using MOAKS). Tissue was then harvested at TKR for BML analysis using histology and tissue microarray.
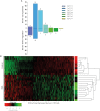
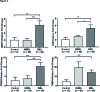
Results: The mean (SD) WOMAC pain scores were significantly increased in advanced OA 59.4 (21.3) and mild OA 30.9 (20.3) compared with controls 0.5 (1.28) (p<0.0001). MOAKS showed all TKR tissue analysed had BMLs, and within these lesions, bone marrow volume was starkly reduced being replaced by dense fibrous connective tissue, new blood vessels, hyaline cartilage and fibrocartilage. Microarray comparing OA BML and normal bone found a significant difference in expression of 218 genes (p<0.05). The most upregulated genes included stathmin 2, thrombospondin 4, matrix metalloproteinase 13 and Wnt/Notch/catenin/chemokine signalling molecules that are known to constitute neuronal, osteogenic and chondrogenic pathways.
Conclusion: Our study is the first to employ detailed histological analysis and microarray techniques to investigate knee OA BMLs. BMLs demonstrated areas of high metabolic activity expressing pain sensitisation, neuronal, extracellular matrix and proinflammatory signalling genes that may explain their strong association with pain.
Keywords: Chemokines; Chondrocytes; Inflammation; Knee Osteoarthritis; Magnetic Resonance Imaging.
© Article author(s) (or their employer(s) unless otherwise stated in the text of the article) 2017. All rights reserved. No commercial use is permitted unless otherwise expressly granted.
Conflict of interest statement
Competing interests: None declared.
Figures


References
-
- Nicholls E, Thomas E, van der Windt DA, et al. Pain trajectory groups in persons with, or at high risk of, knee osteoarthritis: findings from the knee clinical Assessment Study and the Osteoarthritis Initiative. Osteoarthritis Cartilage 2014;22:2041–50. 10.1016/j.joca.2014.09.026 - DOI - PMC - PubMed
-
- Sofat N, Ejindu V, Kiely P. What makes OA painful? The evidence for peripheral and central pain processing Rheumatology. Rheumatology 2011;50:2157–65. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

