A general approach to the design of allosteric, transcription factor-regulated DNAzymes
- PMID: 28706715
- PMCID: PMC5496187
- DOI: 10.1039/c5sc00228a
A general approach to the design of allosteric, transcription factor-regulated DNAzymes
Abstract
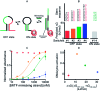
Here we explore a general strategy for the rational design of nucleic acid catalysts that can be allosterically activated by specific nucleic-acid binding proteins. To demonstrate this we have combined a catalytic DNAzyme sequence and the consensus sequence recognized by specific transcription factors to create a construct exhibiting two low-energy conformations: a more stable conformation lacking catalytic activity and lacking the transcription factor binding site, and a less stable conformation that is both catalytically active and competent to bind the transcription factor. The presence of the target transcription factor pushes the equilibrium between these states towards the latter conformation, concomitantly activating catalysis. To demonstrate this we have designed and characterized two peroxidase-like DNAzymes whose activities are triggered upon binding either TATA binding protein or the microphthalmia-associated transcription factor. Our approach augments the current tool kit for the allosteric control of DNAzymes and ribozymes and, because transcription factors control many key biological functions, could have important clinical and diagnostic applications.
Figures
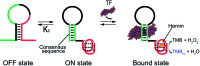
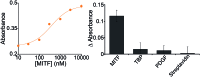

References
-
- Willner I., Shlyahovsky B., Zayats M., Willner B. Chem. Soc. Rev. 2008;37(6):1153–1165. - PubMed
-
- Breaker R. R., Joyce G. F. Trends Biotechnol. 1994;12:268–275. - PubMed
-
- Breaker R. R., Joyce G. F. Chem. Biol. 1994;1(4):223–229. - PubMed
-
- Illangasekare M., Sanchez G., Nickles T., Yarus M. Science. 1995;267(5198):643–647. - PubMed
-
- Jenne A., Famulok M. Chem. Biol. 1998;5(1):23–34. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

