The importance of copy number variation in congenital heart disease
- PMID: 28706735
- PMCID: PMC5505728
- DOI: 10.1038/npjgenmed.2016.31
The importance of copy number variation in congenital heart disease
Abstract
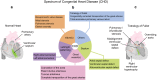
Congenital heart disease (CHD) is the most common class of major malformations in humans. The historical association with large chromosomal abnormalities foreshadowed the role of submicroscopic rare copy number variations (CNVs) as important genetic causes of CHD. Recent studies have provided robust evidence for these structural variants as genome-wide contributors to all forms of CHD, including CHD that appears isolated without extra-cardiac features. Overall, a CNV-related molecular diagnosis can be made in up to one in eight patients with CHD. These include de novo and inherited variants at established (chromosome 22q11.2), emerging (chromosome 1q21.1), and novel loci across the genome. Variable expression of rare CNVs provides support for the notion of a genetic spectrum of CHD that crosses traditional anatomic classification boundaries. Clinical genetic testing using genome-wide technologies (e.g., chromosomal microarray analysis) is increasingly employed in prenatal, paediatric and adult settings. CNV discoveries in CHD have translated to changes to clinical management, prognostication and genetic counselling. The convergence of findings at individual gene and at pathway levels is shedding light on the mechanisms that govern human cardiac morphogenesis. These clinical and research advances are helping to inform whole-genome sequencing, the next logical step in delineating the genetic architecture of CHD.
Conflict of interest statement
COMPETING INTERESTS The authors declare no conflict of interest.
Figures
References
-
- Hoffman, J. I. & Kaplan, S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890–1900 (2002). - PubMed
-
- van der Linde, D. et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J. Am. Coll. Cardiol. 58, 2241–2247 (2011). - PubMed
-
- van der Bom, T. et al. The changing epidemiology of congenital heart disease. Nat. Rev. Cardiol. 8, 50–60 (2011). - PubMed
-
- Friedberg, M. K. et al. Prenatal detection of congenital heart disease. J. Pediatr. 155, 26–31, 31.e1 (2009). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

