The interplay between transcription and mRNA degradation in Saccharomyces cerevisiae
- PMID: 28706937
- PMCID: PMC5507684
- DOI: 10.15698/mic2017.07.580
The interplay between transcription and mRNA degradation in Saccharomyces cerevisiae
Abstract
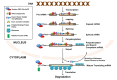
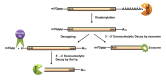
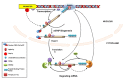
The cellular transcriptome is shaped by both the rates of mRNA synthesis in the nucleus and mRNA degradation in the cytoplasm under a specified condition. The last decade witnessed an exciting development in the field of post-transcriptional regulation of gene expression which underscored a strong functional coupling between the transcription and mRNA degradation. The functional integration is principally mediated by a group of specialized promoters and transcription factors that govern the stability of their cognate transcripts by "marking" them with a specific factor termed "coordinator." The "mark" carried by the message is later decoded in the cytoplasm which involves the stimulation of one or more mRNA-decay factors, either directly by the "coordinator" itself or in an indirect manner. Activation of the decay factor(s), in turn, leads to the alteration of the stability of the marked message in a selective fashion. Thus, the integration between mRNA synthesis and decay plays a potentially significant role to shape appropriate gene expression profiles during cell cycle progression, cell division, cellular differentiation and proliferation, stress, immune and inflammatory responses, and may enhance the rate of biological evolution.
Keywords: CLB2; RPL30; Rpb4/7; SWI5; coordinator; functional coupling; mRNA degradation; mRNA mark; promoter; transcription.
Conflict of interest statement
Conflict of interest: The authors declare no conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
