LINE-1 Hypermethylation in Serum Cell-Free DNA of Relapsing Remitting Multiple Sclerosis Patients
- PMID: 28707075
- PMCID: PMC5948235
- DOI: 10.1007/s12035-017-0679-z
LINE-1 Hypermethylation in Serum Cell-Free DNA of Relapsing Remitting Multiple Sclerosis Patients
Abstract
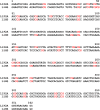
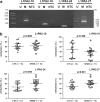
Concentrations of cell-free DNA (cfDNA) circulating in blood and its epigenetic variation, such as DNA methylation, may provide useful diagnostic or prognostic information. Long interspersed nuclear element-1 (LINE-1) constitutes approximately 20% of the human genome and its 5'UTR region is CpG rich. Due to its wide distribution, the methylation level of the 5'UTR of LINE-1 can serve as a surrogate marker of global genomic DNA methylation. The aim of the current study was to investigate whether the methylation status of LINE-1 elements in serum cell-free DNA differs between relapsing remitting multiple sclerosis (RRMS) patients and healthy control subjects (CTR). Serum DNA samples of 6 patients and 6 controls were subjected to bisulfite sequencing. The results showed that the methylation level varies among distinct CpG sites in the 5'UTR of LINE-1 repeats and revealed differences in the methylation state of specific sites in this element between patients and controls. The latter differences were largely due to CpG sites in the L1PA2 subfamily, which were more frequently methylated in the RRMS patients than in the CTR group, whereas such differences were not observed in the L1HS subfamily. These data were verified by quantitative PCR using material from 18 patients and 18 control subjects. The results confirmed that the methylation level of a subset of the CpG sites within the LINE-1 promoter is elevated in DNA from RRMS patients in comparison with CTR. The present data suggest that the methylation status of CpG sites of LINE repeats could be a basis for development of diagnostic or prognostic tests.
Keywords: Cell-free DNA; CpG; DNA methylation; LINE-1; Multiple sclerosis.
Conflict of interest statement
Conflict of Interest
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Long Interspersed Element-1 Methylation Level as a Prognostic Biomarker in Gastrointestinal Cancers.Digestion. 2018;97(1):26-30. doi: 10.1159/000484104. Epub 2018 Feb 1. Digestion. 2018. PMID: 29393154 Review.
-
CpG Island Methylation Patterns in Relapsing-Remitting Multiple Sclerosis.J Mol Neurosci. 2018 Mar;64(3):478-484. doi: 10.1007/s12031-018-1046-x. Epub 2018 Mar 7. J Mol Neurosci. 2018. PMID: 29516350
-
Different genome-wide DNA methylation patterns in CD4+ T lymphocytes and CD14+ monocytes characterize relapse and remission of multiple sclerosis: Focus on GNAS.Mult Scler Relat Disord. 2024 Nov;91:105910. doi: 10.1016/j.msard.2024.105910. Epub 2024 Sep 29. Mult Scler Relat Disord. 2024. PMID: 39369632
-
Methylation patterns of cell-free plasma DNA in relapsing-remitting multiple sclerosis.J Neurol Sci. 2010 Mar 15;290(1-2):16-21. doi: 10.1016/j.jns.2009.12.018. Epub 2010 Jan 12. J Neurol Sci. 2010. PMID: 20064646 Free PMC article.
-
Clinical implications of the LINE-1 methylation levels in patients with gastrointestinal cancer.Surg Today. 2014 Oct;44(10):1807-16. doi: 10.1007/s00595-013-0763-6. Epub 2013 Oct 23. Surg Today. 2014. PMID: 24150097 Review.
Cited by
-
Circulating nucleic acids in the plasma and serum as potential biomarkers in neurological disorders.Braz J Med Biol Res. 2020;53(10):e9881. doi: 10.1590/1414-431x20209881. Epub 2020 Aug 17. Braz J Med Biol Res. 2020. PMID: 32813850 Free PMC article. Review.
-
DNA Methylation As an Epigenetic Mechanism in the Development of Multiple Sclerosis.Acta Naturae. 2021 Apr-Jun;13(2):45-57. doi: 10.32607/actanaturae.11043. Acta Naturae. 2021. PMID: 34377555 Free PMC article.
-
Cell-free DNA-based liquid biopsies in neurology.Brain. 2023 May 2;146(5):1758-1774. doi: 10.1093/brain/awac438. Brain. 2023. PMID: 36408894 Free PMC article. Review.
-
Changes in Deoxyribonucleic Acid Methylation Contribute to the Pathophysiology of Multiple Sclerosis.Front Genet. 2019 Nov 12;10:1138. doi: 10.3389/fgene.2019.01138. eCollection 2019. Front Genet. 2019. PMID: 31798633 Free PMC article. Review.
-
Cell-free DNA methylation as a potential biomarker in brain disorders.Epigenomics. 2022 Apr;14(7):369-374. doi: 10.2217/epi-2021-0416. Epub 2022 Jan 17. Epigenomics. 2022. PMID: 35034473 Free PMC article. No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

