Heterogeneous Enhancement Patterns of Tumor-adjacent Parenchyma at MR Imaging Are Associated with Dysregulated Signaling Pathways and Poor Survival in Breast Cancer
- PMID: 28708462
- PMCID: PMC5673053
- DOI: 10.1148/radiol.2017162823
Heterogeneous Enhancement Patterns of Tumor-adjacent Parenchyma at MR Imaging Are Associated with Dysregulated Signaling Pathways and Poor Survival in Breast Cancer
Abstract
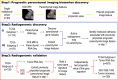
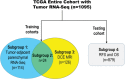
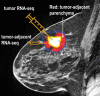
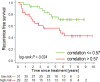
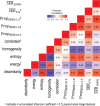
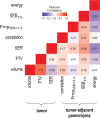
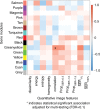
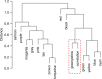
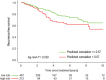
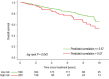
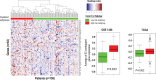
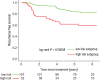
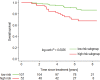
Purpose To identify the molecular basis of quantitative imaging characteristics of tumor-adjacent parenchyma at dynamic contrast material-enhanced magnetic resonance (MR) imaging and to evaluate their prognostic value in breast cancer. Materials and Methods In this institutional review board-approved, HIPAA-compliant study, 10 quantitative imaging features depicting tumor-adjacent parenchymal enhancement patterns were extracted and screened for prognostic features in a discovery cohort of 60 patients. By using data from The Cancer Genome Atlas (TCGA), a radiogenomic map for the tumor-adjacent parenchymal tissue was created and molecular pathways associated with prognostic parenchymal imaging features were identified. Furthermore, a multigene signature of the parenchymal imaging feature was built in a training cohort (n = 126), and its prognostic relevance was evaluated in two independent cohorts (n = 879 and 159). Results One image feature measuring heterogeneity (ie, information measure of correlation) was significantly associated with prognosis (false-discovery rate < 0.1), and at a cutoff of 0.57 stratified patients into two groups with different recurrence-free survival rates (log-rank P = .024). The tumor necrosis factor signaling pathway was identified as the top enriched pathway (hypergeometric P < .0001) among genes associated with the image feature. A 73-gene signature based on the tumor profiles in TCGA achieved good association with the tumor-adjacent parenchymal image feature (R2 = 0.873), which stratified patients into groups regarding recurrence-free survival (log-rank P = .029) and overall survival (log-rank P = .042) in an independent TCGA cohort. The prognostic value was confirmed in another independent cohort (Gene Expression Omnibus GSE 1456), with log-rank P = .00058 for recurrence-free survival and log-rank P = .0026 for overall survival. Conclusion Heterogeneous enhancement patterns of tumor-adjacent parenchyma at MR imaging are associated with the tumor necrosis signaling pathway and poor survival in breast cancer. © RSNA, 2017 Online supplemental material is available for this article.
Figures

Similar articles
-
Tumour heterogeneity revealed by unsupervised decomposition of dynamic contrast-enhanced magnetic resonance imaging is associated with underlying gene expression patterns and poor survival in breast cancer patients.Breast Cancer Res. 2019 Oct 17;21(1):112. doi: 10.1186/s13058-019-1199-8. Breast Cancer Res. 2019. PMID: 31623683 Free PMC article.
-
Magnetic resonance imaging and molecular features associated with tumor-infiltrating lymphocytes in breast cancer.Breast Cancer Res. 2018 Sep 3;20(1):101. doi: 10.1186/s13058-018-1039-2. Breast Cancer Res. 2018. PMID: 30176944 Free PMC article.
-
Unsupervised Clustering of Quantitative Image Phenotypes Reveals Breast Cancer Subtypes with Distinct Prognoses and Molecular Pathways.Clin Cancer Res. 2017 Jul 1;23(13):3334-3342. doi: 10.1158/1078-0432.CCR-16-2415. Epub 2017 Jan 10. Clin Cancer Res. 2017. PMID: 28073839 Free PMC article.
-
Role of texture analysis in breast MRI as a cancer biomarker: A review.J Magn Reson Imaging. 2019 Apr;49(4):927-938. doi: 10.1002/jmri.26556. Epub 2018 Nov 3. J Magn Reson Imaging. 2019. PMID: 30390383 Free PMC article. Review.
-
Imaging genomics for accurate diagnosis and treatment of tumors: A cutting edge overview.Biomed Pharmacother. 2021 Mar;135:111173. doi: 10.1016/j.biopha.2020.111173. Epub 2020 Dec 28. Biomed Pharmacother. 2021. PMID: 33383370 Review.
Cited by
-
Radiomic biomarkers of tumor immune biology and immunotherapy response.Clin Transl Radiat Oncol. 2021 Apr 7;28:97-115. doi: 10.1016/j.ctro.2021.03.006. eCollection 2021 May. Clin Transl Radiat Oncol. 2021. PMID: 33937530 Free PMC article. Review.
-
Contralateral parenchymal enhancement on dynamic contrast-enhanced MRI reproduces as a biomarker of survival in ER-positive/HER2-negative breast cancer patients.Eur Radiol. 2018 Nov;28(11):4705-4716. doi: 10.1007/s00330-018-5470-7. Epub 2018 May 7. Eur Radiol. 2018. PMID: 29736850 Free PMC article.
-
Combination of Peri-Tumoral and Intra-Tumoral Radiomic Features on Bi-Parametric MRI Accurately Stratifies Prostate Cancer Risk: A Multi-Site Study.Cancers (Basel). 2020 Aug 6;12(8):2200. doi: 10.3390/cancers12082200. Cancers (Basel). 2020. PMID: 32781640 Free PMC article.
-
18F-FDG-PET-based radiomics features to distinguish primary central nervous system lymphoma from glioblastoma.Neuroimage Clin. 2019;23:101912. doi: 10.1016/j.nicl.2019.101912. Epub 2019 Jun 27. Neuroimage Clin. 2019. PMID: 31491820 Free PMC article.
-
Prognostic Significance of CT-Attenuation of Tumor-Adjacent Breast Adipose Tissue in Breast Cancer Patients with Surgical Resection.Cancers (Basel). 2019 Aug 8;11(8):1135. doi: 10.3390/cancers11081135. Cancers (Basel). 2019. PMID: 31398863 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

