Modeling polymorphic transformation of rotating bacterial flagella in a viscous fluid
- PMID: 28709256
- PMCID: PMC5656015
- DOI: 10.1103/PhysRevE.95.063106
Modeling polymorphic transformation of rotating bacterial flagella in a viscous fluid
Abstract
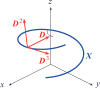
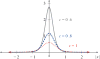
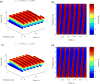
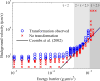
The helical flagella that are attached to the cell body of bacteria such as Escherichia coli and Salmonella typhimurium allow the cell to swim in a fluid environment. These flagella are capable of polymorphic transformation in that they take on various helical shapes that differ in helical pitch, radius, and chirality. We present a mathematical model of a single flagellum described by Kirchhoff rod theory that is immersed in a fluid governed by Stokes equations. We perform numerical simulations to demonstrate two mechanisms by which polymorphic transformation can occur, as observed in experiments. First, we consider a flagellar filament attached to a rotary motor in which transformations are triggered by a reversal of the direction of motor rotation [L. Turner et al., J. Bacteriol. 182, 2793 (2000)10.1128/JB.182.10.2793-2801.2000]. We then consider a filament that is fixed on one end and immersed in an external fluid flow [H. Hotani, J. Mol. Biol. 156, 791 (1982)10.1016/0022-2836(82)90142-5]. The detailed dynamics of the helical flagellum interacting with a viscous fluid is discussed and comparisons with experimental and theoretical results are provided.
Figures








Similar articles
-
Instabilities of a rotating helical rod in a viscous fluid.Phys Rev E. 2017 Feb;95(2-1):022410. doi: 10.1103/PhysRevE.95.022410. Epub 2017 Feb 21. Phys Rev E. 2017. PMID: 28297972
-
Fluid-mechanical interaction of flexible bacterial flagella by the immersed boundary method.Phys Rev E Stat Nonlin Soft Matter Phys. 2012 Mar;85(3 Pt 2):036307. doi: 10.1103/PhysRevE.85.036307. Epub 2012 Mar 19. Phys Rev E Stat Nonlin Soft Matter Phys. 2012. PMID: 22587180
-
Periodic chirality transformations propagating on bacterial flagella.Phys Rev Lett. 2002 Sep 9;89(11):118102. doi: 10.1103/PhysRevLett.89.118102. Epub 2002 Aug 23. Phys Rev Lett. 2002. PMID: 12225172
-
[Mechanism of bacterial motility].Nihon Saikingaku Zasshi. 2019;74(2):157-165. doi: 10.3412/jsb.74.157. Nihon Saikingaku Zasshi. 2019. PMID: 31474648 Review. Japanese.
-
Flagella-Driven Motility of Bacteria.Biomolecules. 2019 Jul 14;9(7):279. doi: 10.3390/biom9070279. Biomolecules. 2019. PMID: 31337100 Free PMC article. Review.
Cited by
-
The swimming of a deforming helix.Eur Phys J E Soft Matter. 2018 Oct 11;41(10):119. doi: 10.1140/epje/i2018-11728-2. Eur Phys J E Soft Matter. 2018. PMID: 30302671
-
Decoding Bacterial Motility: From Swimming States to Patterns and Chemotactic Strategies.Biomolecules. 2025 Jan 23;15(2):170. doi: 10.3390/biom15020170. Biomolecules. 2025. PMID: 40001473 Free PMC article. Review.
-
Modeling and simulation of complex dynamic musculoskeletal architectures.Nat Commun. 2019 Oct 23;10(1):4825. doi: 10.1038/s41467-019-12759-5. Nat Commun. 2019. PMID: 31645555 Free PMC article.
-
Forward and inverse problems in the mechanics of soft filaments.R Soc Open Sci. 2018 Jun 13;5(6):171628. doi: 10.1098/rsos.171628. eCollection 2018 Jun. R Soc Open Sci. 2018. PMID: 30110439 Free PMC article.
-
Architecture and Assembly of the Bacterial Flagellar Motor Complex.Subcell Biochem. 2021;96:297-321. doi: 10.1007/978-3-030-58971-4_8. Subcell Biochem. 2021. PMID: 33252734 Review.
References
-
- Lauga E. Annu. Rev. Fluid Mech. 2016;48:105.
-
- Li C, Motaleb A, Sal M, Goldstein SF, Charon NW. J. Mol. Microbiol. Biotechnol. 2000;2:345. - PubMed
-
- McBride MJ. Annu. Rev. Microbiol. 2001;55:49. - PubMed
-
- Weibull C. In: The Bacteria: A Treatise on Structure and Function. Gunsalus IC, Stanier RY, editors. Vol. 1. Academic Press; New York: 1960. pp. 153–205.
-
- Berg HC. Annu. Rev. Biochem. 2003;72:19. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
