Clonal competition in BcrAbl-driven leukemia: how transplantations can accelerate clonal conversion
- PMID: 28709463
- PMCID: PMC5512731
- DOI: 10.1186/s12943-017-0668-x
Clonal competition in BcrAbl-driven leukemia: how transplantations can accelerate clonal conversion
Abstract
Background: Clonal competition in cancer describes the process in which the progeny of a cell clone supersedes or succumbs to other competing clones due to differences in their functional characteristics, mostly based on subsequently acquired mutations. Even though the patterns of those mutations are well explored in many tumors, the dynamical process of clonal selection is underexposed.
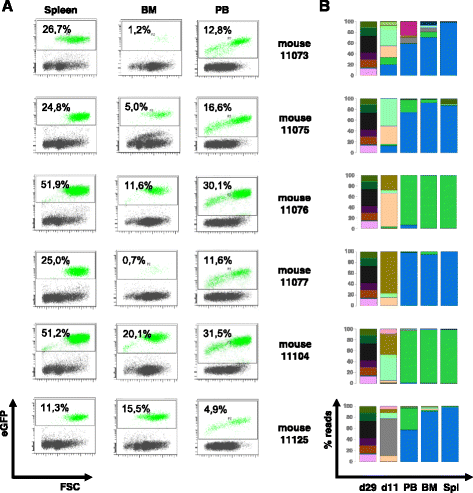
Methods: We studied the dynamics of clonal competition in a BcrAbl-induced leukemia using a γ-retroviral vector library encoding the oncogene in conjunction with genetic barcodes. To this end, we studied the growth dynamics of transduced cells on the clonal level both in vitro and in vivo in transplanted mice.
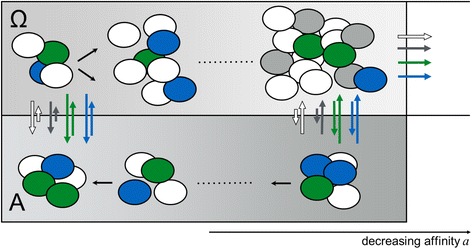
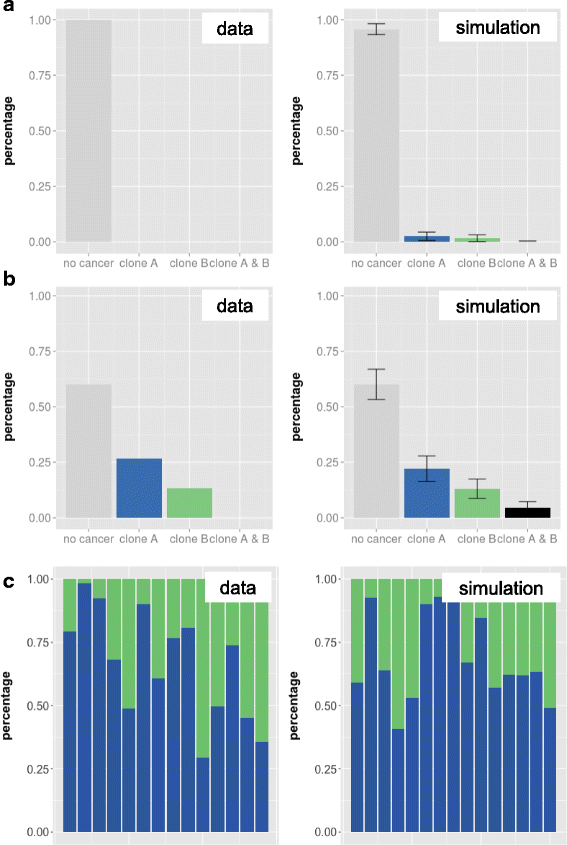
Results: While we detected moderate changes in clonal abundancies in vitro, we observed monoclonal leukemias in 6/30 mice after transplantation, which intriguingly were caused by only two different BcrAbl clones. To analyze the success of these clones, we applied a mathematical model of hematopoietic tissue maintenance, which indicated that a differential engraftment capacity of these two dominant clones provides a possible explanation of our observations. These findings were further supported by additional transplantation experiments and increased BcrAbl transcript levels in both clones.
Conclusion: Our findings show that clonal competition is not an absolute process based on mutations, but highly dependent on selection mechanisms in a given environmental context.
Keywords: BcrAbl; Clonal competition; Clonal dynamics; Genetic barcodes; Heterogeneity; Leukemia; Mathematical modelling.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

