Mechanism and properties of positive allosteric modulation of N-methyl-d-aspartate receptors by 6-alkyl 2-naphthoic acid derivatives
- PMID: 28709671
- PMCID: PMC5623084
- DOI: 10.1016/j.neuropharm.2017.07.007
Mechanism and properties of positive allosteric modulation of N-methyl-d-aspartate receptors by 6-alkyl 2-naphthoic acid derivatives
Abstract
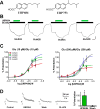
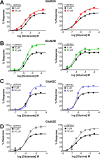
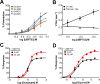
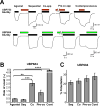
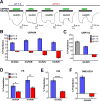
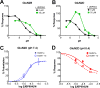
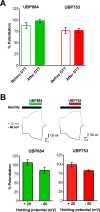
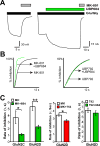
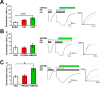
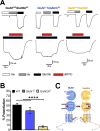
The theory that N-methyl-d-aspartate receptor (NMDAR) hypofunction is responsible for the symptoms of schizophrenia is well supported by many pharmacological and genetic studies. Accordingly, positive allosteric modulators (PAMs) that augment NMDAR signaling may be useful for treating schizophrenia. Previously we have identified several NMDAR PAMs containing a carboxylic acid attached to naphthalene, phenanthrene, or coumarin ring systems. In this study, we describe several functional and mechanistic properties of UBP684, a 2-naphthoic acid derivative, which robustly potentiates agonist responses at each of the four GluN1a/GluN2 receptors and at neuronal NMDARs. UBP684 increases the maximal l-glutamate/glycine response while having minor subunit-specific effects on agonist potency. PAM binding is independent of agonist binding, and PAM activity is independent of membrane voltage, redox state, and the GluN1 exon 5 N-terminal insert. UBP684 activity is, however, markedly pH-dependent, with greater potentiation occurring at lower pHs and inhibitory activity at pH 8.4. UBP684 increases channel open probability (Po) and slows receptor deactivation time upon removal of l-glutamate, but not glycine. The structurally related PAM, UBP753, reproduced most of these findings, but did not prolong agonist removal deactivation time. Studies using cysteine mutants to lock the GluN1 and GluN2 ligand-binding domains (LBDs) in the agonist-bound states indicate that PAM potentiation requires GluN2 LBD conformational flexibility. Together, these findings suggest that UBP684 and UBP753 stabilize the GluN2 LBD in an active conformation and thereby increase Po. Thus, UBP684 and UBP753 may serve as lead compounds for developing agents to enhance NMDAR activity in disorders associated with NMDAR hypofunction.
Keywords: Deactivation; Ligand-binding domain; N-methyl-d-aspartate; Positive allosteric modulator; Potentiator; l-glutamate.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
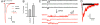
References
-
- Bettini E, Sava A, Griffante C, Carignani C, Buson A, Capelli AM, Negri M, Andreetta F, Senar-Sancho SA, Guiral L, Cardullo F. Identification and characterisation of novel NMDA receptor antagonists selective for NR2A- over NR2B-containing receptors. J Pharmacol Exp Ther. 2010;335:636–644. - PubMed
-
- Ceccon M, Rumbaugh G, Vicini S. Distinct effect of pregnenolone sulfate on NMDA receptor subtypes. Neuropharmacology. 2001;40:491–500. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

