Human Secretory IgM Emerges from Plasma Cells Clonally Related to Gut Memory B Cells and Targets Highly Diverse Commensals
- PMID: 28709802
- PMCID: PMC5519504
- DOI: 10.1016/j.immuni.2017.06.013
Human Secretory IgM Emerges from Plasma Cells Clonally Related to Gut Memory B Cells and Targets Highly Diverse Commensals
Abstract
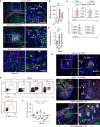
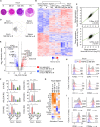
Secretory immunoglobulin A (SIgA) enhances host-microbiota symbiosis, whereas SIgM remains poorly understood. We found that gut IgM+ plasma cells (PCs) were more abundant in humans than mice and clonally related to a large repertoire of memory IgM+ B cells disseminated throughout the intestine but rare in systemic lymphoid organs. In addition to sharing a gut-specific gene signature with memory IgA+ B cells, memory IgM+ B cells were related to some IgA+ clonotypes and switched to IgA in response to T cell-independent or T cell-dependent signals. These signals induced abundant IgM which, together with SIgM from clonally affiliated PCs, recognized mucus-embedded commensals. Bacteria recognized by human SIgM were dually coated by SIgA and showed increased richness and diversity compared to IgA-only-coated or uncoated bacteria. Thus, SIgM may emerge from pre-existing memory rather than newly activated naive IgM+ B cells and could help SIgA to anchor highly diverse commensal communities to mucus.
Keywords: IgA; IgM; gut; human; memory B cells; microbiota; mucosa; plasma cells; repertoire.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Aranburu A., Piano Mortari E., Baban A., Giorda E., Cascioli S., Marcellini V., Scarsella M., Ceccarelli S., Corbelli S., Cantarutti N. Human B-cell memory is shaped by age- and tissue-specific T-independent and GC-dependent events. Eur. J. Immunol. 2017;47:327–344. - PubMed
-
- Barak M., Zuckerman N.S., Edelman H., Unger R., Mehr R. IgTree: creating Immunoglobulin variable region gene lineage trees. J. Immunol. Methods. 2008;338:67–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

