An efficient algorithm for generating the internal branches of a Kingman coalescent
- PMID: 28709926
- PMCID: PMC5764821
- DOI: 10.1016/j.tpb.2017.05.002
An efficient algorithm for generating the internal branches of a Kingman coalescent
Abstract
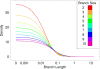
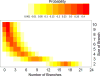
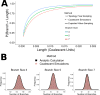
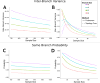
Coalescent simulations are a widely used approach for simulating sample genealogies, but can become computationally burdensome in large samples. Methods exist to analytically calculate a sample's expected frequency spectrum without simulating full genealogies. However, statistics that rely on the distribution of the length of internal coalescent branches, such as the probability that two mutations of equal size arose on the same genealogical branch, have previously required full coalescent simulations to estimate. Here, we present a sampling method capable of efficiently generating limited portions of sample genealogies using a series of analytic equations that give probabilities for the number, start, and end of internal branches conditional on the number of final samples they subtend. These equations are independent of the coalescent waiting times and need only be calculated a single time, lending themselves to efficient computation. We compare our method with full coalescent simulations to show the resulting distribution of branch lengths and summary statistics are equivalent, but that for many conditions our method is at least 10 times faster.
Keywords: Coalescent; Coalescent simulations; Genealogical topology.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures

References
-
- Berndt S, Gustafsson S, Mägi R, Ganna A, Wheeler E, Feitosa M, Justice A, Monda K, Croteau-Chonka D, Day F, Esko T, Fall T, Ferreira T, Gentilini D, Jackson A, Luan J, Randall J, Vedantam S, Willer C, Winkler T, Wood A, Workalemahu T, Hu Y, Lee S, Liang L, Lin D, Min J, Neale B, Thorleifsson G, Yang J, Albrecht E, Amin N, Bragg-Gresham J, Cadby G, den Heijer M, Eklund N, Fischer K, Goel A, Hottenga J, Huffman J, Jarick I, Johansson A, Johnson T, Kanoni S, Kleber M, König I, Kristiansson K, Kutalik Z, Lamina C, Lecoeur C, Li G, Mangino M, McArdle W, Medina-Gomez C, Müller-Nurasyid M, Ngwa J, Nolte I, Paternoster L, Pechlivanis S, Perola M, Peters M, Preuss M, Rose L, Shi J, Shungin D, Smith A, Strawbridge R, Surakka I, Teumer A, Trip M, Tyrer J, Van Vliet-Ostaptchouk J, Vandenput L, Waite L, Zhao J, Absher D, Asselbergs F, Atalay M, Attwood A, Balmforth A, Basart H, Beilby J, Bonnycastle L, Brambilla P, Bruinenberg M, Campbell H, Chasman D, Chines P, Collins F, Connell J, Cookson W, de Faire U, de Vegt F, Dei M, Dimitriou M, Edkins S, Estrada K, Evans D, Farrall M, Ferrario M, Ferriéres J, Franke L, Frau F, Gejman P, Grallert H, Grönberg H, Gudnason V, Hall A, Hall P, Hartikainen A, Hayward C, Heard-Costa N, Heath A, Hebebrand J, Homuth G, Hu F, Hunt S, Hyppönen E, Iribarren C, Jacobs K, Jansson J, Jula A, Kähönen M, Kathiresan S, Kee F, Khaw K, Kivimäki M, Koenig W, Kraja A, Kumari M, Kuulasmaa K, Kuusisto J, Laitinen J, Lakka T, Langenberg C, Launer L, Lind L, Lindström J, Liu J, Liuzzi A, Lokki M, Lorentzon M, Madden P, Magnusson P, Manunta P, Marek D, März W, Mateo Leach I, McKnight B, Medland S, Mihailov E, Milani L, Montgomery GVM, Mühleisen T, Munroe P, Musk A, Narisu N, Navis G, Nicholson G, Nohr E, Ong K, Oostra B, Palmer C, Palotie A, Peden J, Ped-ersen N, Peters A, Polasek O, Pouta A, Pramstaller P, Prokopenko I, Pütter C, Radhakrishnan A, Raitakari O, Rendon A, Rivadeneira F, Rudan I, Saaristo T, Sambrook J, Sanders A, Sanna S, Saramies J, Schipf S, Schreiber S, Schunkert H, Shin S, Signorini S, Sinisalo J, Skrobek B, Soranzo N, Stančáková A, Stark K, Stephens J, Stirrups K, Stolk R, Stumvoll M, Swift A, Theodoraki E, Thorand B, Tregouet D, Tremoli E, Van der Klauw M, van Meurs J, Vermeulen S, Viikari J, Virtamo J, Vitart V, Waeber G, Wang Z, Widèn E, Wild S, Willemsen G, Winkelmann B, Witteman J, Wolffenbuttel B, Wong A, Wright A, Zillikens M, Amouyel P, Boehm B, Boerwinkle E, Boomsma D, Caulfield M, Chanock S, Cupples L, Cusi D, Dedoussis G, Erdmann J, Eriksson J, Franks P, Froguel P, Gieger C, Gyllensten U, Hamsten A, Harris T, Hengstenberg C, Hicks A, Hingorani A, Hinney A, Hofman A, Hovingh K, Hveem K, Illig T, Jarvelin M, Jöckel K, Keinanen-Kiukaanniemi S, Kiemeney L, Kuh D, Laakso M, Lehtimäki T, Levinson D, Martin N, Metspalu A, Morris A, Nieminen M, Njølstad I, Ohlsson C, Oldehinkel A, Ouwehand W, Palmer L, Penninx B, Power C, Province M, Psaty B, Qi L, Rauramaa R, Ridker P, Ripatti S, Salomaa V, Samani N, Snieder H, Sørensen T, Spector T, Stefansson K, Tönjes A, Tuomilehto J, Uitterlinden A, Uusitupa M, van der Harst P, Vollenweider P, Wallaschofski H, Wareham N, Watkins H, Wichmann H, Wilson J, Abecasis G, Assimes T, Barroso I, Boehnke M, Borecki I, Deloukas P, Fox C, Frayling T, Groop L, Haritunian T, Heid I, Hunter D, Kaplan R, Karpe F, Moffatt M, Mohlke K, O’Connell J, Pawitan Y, Schadt E, Schlessinger D, Steinthorsdottir V, Strachan D, Thorsteinsdottir U, Visscher P, Di Blasio A, Hirschhorn J, Lindgren C, Morris A, Meyre D, Scherag A, McCarthy M, Speliotes E, North K, Loos R, Ingelsson E. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–12. - PMC - PubMed
-
- Coventry A, Bull-Otterson L, Liu X, Clark A, Maxwell T, Crosby J, Hixson J, Rea T, Muzny D, Lewis L, Wheeler D, Sabo A, Lusk C, Weiss K, Akbar H, Cree A, Hawes A, Newsham I, Varghese R, Villasana D, Gross S, Joshi V, Santibanez J, Morgan M, Chang K, Hale W, IV, Templeton A, Boerwinkle E, Gibbs R, Sing C. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nature Communications. 2010;1:131. - PMC - PubMed
-
- Dahmer I, Kersting G. The internal branch lengths of the Kingman coalescent. Ann Appl Probab. 2015:1325–1348.
-
- Dhersin J, Mölhe M. On the external branches of coalescents with multiple collisions. Electron J Probab. 2013;18(40):11.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

