Gene Regulatory Networks for the Haploid-to-Diploid Transition of Chlamydomonas reinhardtii
- PMID: 28710131
- PMCID: PMC5580766
- DOI: 10.1104/pp.17.00731
Gene Regulatory Networks for the Haploid-to-Diploid Transition of Chlamydomonas reinhardtii
Abstract
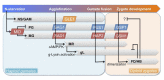
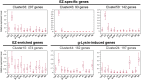
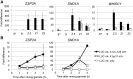
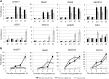
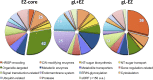
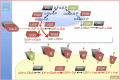
The sexual cycle of the unicellular Chlamydomonas reinhardtii culminates in the formation of diploid zygotes that differentiate into dormant spores that eventually undergo meiosis. Mating between gametes induces rapid cell wall shedding via the enzyme g-lysin; cell fusion is followed by heterodimerization of sex-specific homeobox transcription factors, GSM1 and GSP1, and initiation of zygote-specific gene expression. To investigate the genetic underpinnings of the zygote developmental pathway, we performed comparative transcriptome analysis of both pre- and post-fertilization samples. We identified 253 transcripts specifically enriched in early zygotes, 82% of which were not up-regulated in gsp1 null zygotes. We also found that the GSM1/GSP1 heterodimer negatively regulates the vegetative wall program at the posttranscriptional level, enabling prompt transition from vegetative wall to zygotic wall assembly. Annotation of the g-lysin-induced and early zygote genes reveals distinct vegetative and zygotic wall programs, supported by concerted up-regulation of genes encoding cell wall-modifying enzymes and proteins involved in nucleotide-sugar metabolism. The haploid-to-diploid transition in Chlamydomonas is masterfully controlled by the GSM1/GSP1 heterodimer, translating fertilization and gamete coalescence into a bona fide differentiation program. The fertilization-triggered integration of genes required to make related, but structurally and functionally distinct organelles-the vegetative versus zygote cell wall-presents a likely scenario for the evolution of complex developmental gene regulatory networks.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures

References
-
- Aoyama H, Hagiwara Y, Misumi O, Kuroiwa T, Nakamura S (2006) Complete elimination of maternal mitochondrial DNA during meiosis resulting in the paternal inheritance of the mitochondrial genome in Chlamydomonas species. Protoplasma 228: 231–242 - PubMed
-
- Atmodjo MA, Hao Z, and Mohnen D (2013) Evolving Views of Pectin Biosynthesis. Annu Rev Plant Biol 64: 747–779 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

