Phosphorylation of human aquaporin 2 (AQP2) allosterically controls its interaction with the lysosomal trafficking protein LIP5
- PMID: 28710278
- PMCID: PMC5582854
- DOI: 10.1074/jbc.M117.788364
Phosphorylation of human aquaporin 2 (AQP2) allosterically controls its interaction with the lysosomal trafficking protein LIP5
Abstract
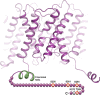
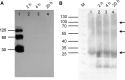
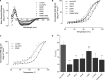
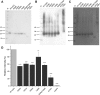
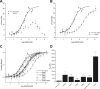
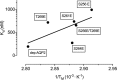
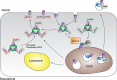
The interaction between the renal water channel aquaporin-2 (AQP2) and the lysosomal trafficking regulator-interacting protein LIP5 targets AQP2 to multivesicular bodies and facilitates lysosomal degradation. This interaction is part of a process that controls AQP2 apical membrane abundance in a vasopressin-dependent manner, allowing for urine volume adjustment. Vasopressin regulates phosphorylation at four sites within the AQP2 C terminus (Ser256, Ser261, Ser264, and Thr269), of which Ser256 is crucial and sufficient for AQP2 translocation from storage vesicles to the apical membrane. However, whether AQP2 phosphorylation modulates AQP2-LIP5 complex affinity is unknown. Here we used far-Western blot analysis and microscale thermophoresis to show that the AQP2 binds LIP5 in a phosphorylation-dependent manner. We constructed five phospho-mimicking mutants (S256E, S261E, S264E, T269E, and S256E/T269E) and a C-terminal truncation mutant (ΔP242) that lacked all phosphorylation sites but retained a previously suggested LIP5-binding site. CD spectroscopy indicated that wild-type AQP2 and the phospho-mimicking mutants had similar overall structure but displayed differences in melting temperatures possibly arising from C-terminal conformational changes. Non-phosphorylated AQP2 bound LIP5 with the highest affinity, whereas AQP2-ΔP242 had 20-fold lower affinity as determined by microscale thermophoresis. AQP2-S256E, S261E, T269E, and S256E/T269E all had reduced affinity. This effect was most prominent for AQP2-S256E, which fits well with its role in apical membrane targeting. AQP2-S264E had affinity similar to non-phosphorylated AQP2, possibly indicating a role in exosome excretion. Our data suggest that AQP2 phosphorylation allosterically controls its interaction with LIP5, illustrating how altered affinities to interacting proteins form the basis for regulation of AQP2 trafficking by post-translational modifications.
Keywords: aquaporin; membrane protein; membrane trafficking; phosphorylation; protein-protein interaction.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
References
-
- Fushimi K., Sasaki S., and Marumo F. (1997) Phosphorylation of serine 256 is required for cAMP-dependent regulatory exocytosis of the aquaporin-2 water channel. J. Biol. Chem. 272, 14800–14804 - PubMed
-
- Katsura T., Gustafson C. E., Ausiello D. A., and Brown D. (1997) Protein kinase A phosphorylation is involved in regulated exocytosis of aquaporin-2 in transfected LLC-PK1 cells. Am. J. Physiol. 272, F817–F822 - PubMed
-
- van Balkom B. W., Savelkoul P. J., Markovich D., Hofman E., Nielsen S., van der Sluijs P., and Deen P. M. (2002) The role of putative phosphorylation sites in the targeting and shuttling of the aquaporin-2 water channel. J. Biol. Chem. 277, 41473–41479 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

