Relationships between gastrointestinal microbiota and blood group antigens
- PMID: 28710295
- PMCID: PMC5625272
- DOI: 10.1152/physiolgenomics.00043.2017
Relationships between gastrointestinal microbiota and blood group antigens
Abstract
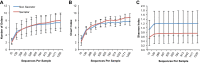
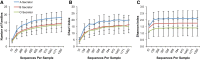
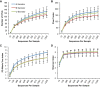
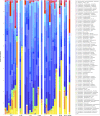
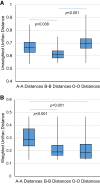
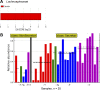
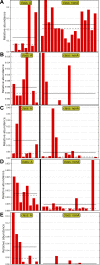
FUT2 is a gene for a fucosyltransferase that encodes expression of ABO blood group antigens found on gastrointestinal mucosa and secretions. We hypothesized that the fecal microbiomes of healthy subjects, with blood group antigens A, B, and O, have differing compositions. We analyzed 33 fecal and blood specimens from healthy subjects for FUT2 genotype, and the fecal microbiome was determined by 454 pyrosequencing. Our data show that being a blood group secretor is associated with less diversity at higher orders of taxonomy; and the presence of blood group A antigens in the secretor subjects are associated with an expansion families of bacteria within the gut. Furthermore, our study confirms the previous findings that secretors and nonsecretors have differing bacterial taxa. This extends the previous findings by demonstrating that the impact of being a nonsecretor is higher than that of individual blood group antigens. Additionally, we demonstrate that both secretor status and blood group antigen expression especially affect the Lachnospiraceae family of bacteria within the gut microbiome, with lower abundances noted in nonsecretors and higher abundances in secretors of various blood groups. We further note specific differences in blood group A-secretors demonstrating that the genus Blautia is lower in the group A-secretors compared with the non-A-secretors and that this reduction is accompanied by higher abundances of members of the Rikenellaceae, Peptostreptococcaceae, Clostridiales, and Turicibacter This study offers a first insight into the relationship between the fecal microbiome and blood group antigens in secretors.
Keywords: bacteria; blood group antigens; fucosyltransferase- FUT2; gastrointestinal microbiome; gastrointestinal secretions.
Copyright © 2017 the American Physiological Society.
Figures

References
-
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M’rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P; MetaHIT Consortium . Enterotypes of the human gut microbiome. Nature 473: 174–180, 2011. doi: 10.1038/nature09944. - DOI - PMC - PubMed
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336, 2010. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

