Persistent circulation of Coxsackievirus A6 of genotype D3 in mainland of China between 2008 and 2015
- PMID: 28710474
- PMCID: PMC5511160
- DOI: 10.1038/s41598-017-05618-0
Persistent circulation of Coxsackievirus A6 of genotype D3 in mainland of China between 2008 and 2015
Abstract
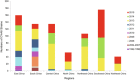
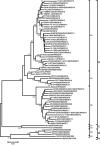
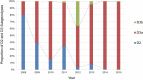
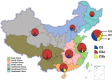
A total of 807 entire VP1 sequences of Coxsackievirus A6 (CV-A6) from mainland of China from 1992 to 2015, including 520 in this study and 287 from the GenBank database, were analysed to provide a basic framework of molecular epidemiological characteristics of CV-A6 in China. Sixty-five VP1 sequences including 46 representative CV-A6 isolates from 807 Chinese strains and 19 international strains from GenBank were used for describing the genotypes and sub-genotypes. The results revealed that CV-A6 strains can be categorised into 4 genotypes designated as A, B, C, and D according to previous data and can be further subdivided into B1-B2, C1-C2, and D1-D3 sub-genotypes. D3 is the predominant sub-genotype that circulated in recent years in mainland of China and represents 734 of 807 Chinese isolates. Sixty-six strains belong to D2, whereas B1 and C1 comprise a single strain each, and five AFP strains formed B2. Sub-genotype D3 first circulated in 2008 and has become the predominant sub-genotype since 2009 and then reached a peak in 2013, while D2 was mostly undetectable in the past years. These data revealed different transmission stages of CV-A6 in mainland of China and that sub-genotype D3 may have stronger transmission ability.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Picornaviridae. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. (Elsevier, San Diego, 2011).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

