UCSF ChimeraX: Meeting modern challenges in visualization and analysis
- PMID: 28710774
- PMCID: PMC5734306
- DOI: 10.1002/pro.3235
UCSF ChimeraX: Meeting modern challenges in visualization and analysis
Abstract
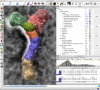
UCSF ChimeraX is next-generation software for the visualization and analysis of molecular structures, density maps, 3D microscopy, and associated data. It addresses challenges in the size, scope, and disparate types of data attendant with cutting-edge experimental methods, while providing advanced options for high-quality rendering (interactive ambient occlusion, reliable molecular surface calculations, etc.) and professional approaches to software design and distribution. This article highlights some specific advances in the areas of visualization and usability, performance, and extensibility. ChimeraX is free for noncommercial use and is available from http://www.rbvi.ucsf.edu/chimerax/ for Windows, Mac, and Linux.
Keywords: biomolecular visualization and analysis; density maps; integrative hybrid modeling; interactive molecular graphics; large-scale data; light-sheet microscopy; structural biology; virtual reality.
© 2017 The Protein Society.
Figures




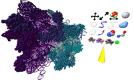
References
-
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, 3rd , Liu Z, English BP, Mimori‐Kiyosue Y, Romero DP, Ritter AT, Lippincott‐Schwartz J, Fritz‐Laylin L, Mullins RD, Mitchell DM, Bembenek JN, Reymann AC, Bohme R, Grill SW, Wang JT, Seydoux G, Tulu US, Kiehart DP, Betzig E (2014) Lattice light‐sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346:1257998. - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

