The Gut Microbiota of Healthy Chilean Subjects Reveals a High Abundance of the Phylum Verrucomicrobia
- PMID: 28713349
- PMCID: PMC5491548
- DOI: 10.3389/fmicb.2017.01221
The Gut Microbiota of Healthy Chilean Subjects Reveals a High Abundance of the Phylum Verrucomicrobia
Abstract
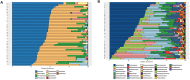
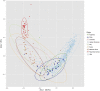
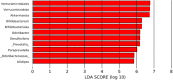
The gut microbiota is currently recognized as an important factor regulating the homeostasis of the gastrointestinal tract and influencing the energetic metabolism of the host as well as its immune and central nervous systems. Determining the gut microbiota composition of healthy subjects is therefore necessary to establish a baseline allowing the detection of microbiota alterations in pathologic conditions. Accordingly, the aim of this study was to characterize the gut microbiota of healthy Chilean subjects using 16S rRNA gene sequencing. Fecal samples were collected from 41 young, asymptomatic, normal weight volunteers (age: 25 ± 4 years; ♀:48.8%; BMI: 22.5 ± 1.6 kg/m2) with low levels of plasma (IL6 and hsCRP) and colonic (fecal calprotectin) inflammatory markers. The V3-V4 region of the 16S rRNA gene of bacterial DNA was amplified and sequenced using MiSeq Illumina system. 109,180 ± 13,148 sequences/sample were obtained, with an α-diversity of 3.86 ± 0.37. The dominant phyla were Firmicutes (43.6 ± 9.2%) and Bacteroidetes (41.6 ± 13.1%), followed by Verrucomicrobia (8.5 ± 10.4%), Proteobacteria (2.8 ± 4.8%), Actinobacteria (1.8 ± 3.9%) and Euryarchaeota (1.4 ± 2.7%). The core microbiota representing the genera present in all the subjects included Bacteroides, Prevotella, Parabacteroides (phylum Bacteroidetes), Phascolarctobacterium, Faecalibacterium, Ruminococcus, Lachnospira, Oscillospira, Blautia, Dorea, Roseburia, Coprococcus, Clostridium, Streptococcus (phylum Firmicutes), Akkermansia (phylum Verrucomicrobia), and Collinsella (phylum Actinobacteria). Butyrate-producing genera including Faecalibacterium, Roseburia, Coprococcus, and Oscillospira were detected. The family Methanobacteriaceae was reported in 83% of the subjects and Desulfovibrio, the most representative sulfate-reducing genus, in 76%. The microbiota of the Chilean individuals significantly differed from those of Papua New Guinea and the Matses ethnic group and was closer to that of the Argentinians and sub-populations from the United States. Interestingly, the microbiota of the Chilean subjects stands out for its richness in Verrucomicrobia; the mucus-degrading bacterium Akkermansia muciniphila is the only identified member of this phylum. This is an important finding considering that this microorganism has been recently proposed as a hallmark of healthy gut due to its anti-inflammatory and immunostimulant properties and its ability to improve gut barrier function, insulin sensitivity and endotoxinemia. These results constitute an important baseline that will facilitate the characterization of dysbiosis in the main diseases affecting the Chilean population.
Keywords: 16S rRNA gene sequencing; Chilean gut microbiota; bacterial communities; fecal samples; healthy normal weight.
Figures

References
-
- Anhê F. F., Roy D., Pilon G., Dudonné S., Matamoros S., Varin T. V., et al. (2015). A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 64 872–883. 10.1136/gutjnl-2014-307142 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

