System-Level and Granger Network Analysis of Integrated Proteomic and Metabolomic Dynamics Identifies Key Points of Grape Berry Development at the Interface of Primary and Secondary Metabolism
- PMID: 28713396
- PMCID: PMC5491621
- DOI: 10.3389/fpls.2017.01066
System-Level and Granger Network Analysis of Integrated Proteomic and Metabolomic Dynamics Identifies Key Points of Grape Berry Development at the Interface of Primary and Secondary Metabolism
Abstract
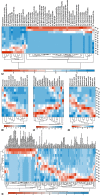
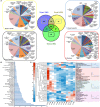
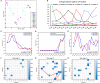
Grapevine is a fruit crop with worldwide economic importance. The grape berry undergoes complex biochemical changes from fruit set until ripening. This ripening process and production processes define the wine quality. Thus, a thorough understanding of berry ripening is crucial for the prediction of wine quality. For a systemic analysis of grape berry development we applied mass spectrometry based platforms to analyse the metabolome and proteome of Early Campbell at 12 stages covering major developmental phases. Primary metabolites involved in central carbon metabolism, such as sugars, organic acids and amino acids together with various bioactive secondary metabolites like flavonols, flavan-3-ols and anthocyanins were annotated and quantified. At the same time, the proteomic analysis revealed the protein dynamics of the developing grape berries. Multivariate statistical analysis of the integrated metabolomic and proteomic dataset revealed the growth trajectory and corresponding metabolites and proteins contributing most to the specific developmental process. K-means clustering analysis revealed 12 highly specific clusters of co-regulated metabolites and proteins. Granger causality network analysis allowed for the identification of time-shift correlations between metabolite-metabolite, protein- protein and protein-metabolite pairs which is especially interesting for the understanding of developmental processes. The integration of metabolite and protein dynamics with their corresponding biochemical pathways revealed an energy-linked metabolism before veraison with high abundances of amino acids and accumulation of organic acids, followed by protein and secondary metabolite synthesis. Anthocyanins were strongly accumulated after veraison whereas other flavonoids were in higher abundance at early developmental stages and decreased during the grape berry developmental processes. A comparison of the anthocyanin profile of Early Campbell to other cultivars revealed similarities to Concord grape and indicates the strong effect of genetic background on metabolic partitioning in primary and secondary metabolism.
Keywords: Vitis vinifera; berry development; data integration; flavonoids; mass spectrometry; primary metabolism; secondary metabolism; systems biology.
Figures


References
-
- Adisakwattana S., Moonrat J., Srichairat S., Chanasit C., Tirapongporn H., Chanathong B., et al. (2010). Lipid-Lowering mechanisms of grape seed extract (Vitis vinifera L.) and its antihyperlidemic activity. J. Med. Plants Res. 4, 2113–2120.
-
- Agudelo-Romero P., Erban A., Sousa L., Pais M. S., Kopka J., Fortes A. M. (2013). Search for transcriptional and metabolic markers of grape pre-ripening and ripening and insights into specific aroma development in three Portuguese cultivars. PLoS ONE 8:e60422. 10.1371/journal.pone.0060422 - DOI - PMC - PubMed
-
- Aharoni A., Keizer L. C., Van Den Broeck H. C., Blanco-Portales R., Munoz-Blanco J., Bois G., et al. (2002). Novel insight into vascular, stress, and auxin-dependent and -independent gene expression programs in strawberry, a non-climacteric fruit. Plant Physiol. 129, 1019–1031. 10.1104/pp.003558 - DOI - PMC - PubMed
-
- Ahn S. Y., Kim S. A., Choi S. J., Yun H. K. (2015). Comparison of accumulation of stilbene compounds and stilbene related gene expression in two grape berries irradiated with different light sources. Hortic. Environ. Biotechnol. 56, 36–43. 10.1007/s13580-015-0045-x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

