Genomic landscape of high-grade meningiomas
- PMID: 28713588
- PMCID: PMC5506858
- DOI: 10.1038/s41525-017-0014-7
Genomic landscape of high-grade meningiomas
Erratum in
-
Erratum: Genomic landscape of high-grade meningiomas.NPJ Genom Med. 2017 Sep 4;2:26. doi: 10.1038/s41525-017-0023-6. eCollection 2017. NPJ Genom Med. 2017. PMID: 29263836 Free PMC article.
Abstract
High-grade meningiomas frequently recur and are associated with high rates of morbidity and mortality. To determine the factors that promote the development and evolution of these tumors, we analyzed the genomes of 134 high-grade meningiomas and compared this information with data from 587 previously published meningiomas. High-grade meningiomas had a higher mutation burden than low-grade meningiomas but did not harbor any statistically significant mutated genes aside from NF2. High-grade meningiomas also possessed significantly elevated rates of chromosomal gains and losses, especially among tumors with monosomy 22. Meningiomas previously treated with adjuvant radiation had significantly more copy number alterations than radiation-induced or radiation-naïve meningiomas. Across serial recurrences, genomic disruption preceded the emergence of nearly all mutations, remained largely uniform across time, and when present in low-grade meningiomas, correlated with subsequent progression to a higher grade. In contrast to the largely stable copy number alterations, mutations were strikingly heterogeneous across tumor recurrences, likely due to extensive geographic heterogeneity in the primary tumor. While high-grade meningiomas harbored significantly fewer overtly targetable alterations than low-grade meningiomas, they contained numerous mutations that are predicted to be neoantigens, suggesting that immunologic targeting may be of therapeutic value.
Conflict of interest statement
The authors declare no competing interests.
Figures
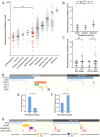


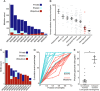
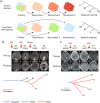
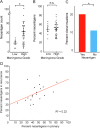
References
-
- Louis, D., Ohgaki, H., Wiestler, O. D. & Cavenee, W. K. WHO Classification of Tumours of the Central Nervous System, 408 (International Agency for Research on Cancer, 2016).
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

