The role of de novo mutations in the development of amyotrophic lateral sclerosis
- PMID: 28714244
- PMCID: PMC6599399
- DOI: 10.1002/humu.23295
The role of de novo mutations in the development of amyotrophic lateral sclerosis
Abstract
The genetic basis combined with the sporadic occurrence of amyotrophic lateral sclerosis (ALS) suggests a role of de novo mutations in disease pathogenesis. Previous studies provided some evidence for this hypothesis; however, results were conflicting: no genes with recurrent occurring de novo mutations were identified and different pathways were postulated. In this study, we analyzed whole-exome data from 82 new patient-parents trios and combined it with the datasets of all previously published ALS trios (173 trios in total). The per patient de novo rate was not higher than expected based on the general population (P = 0.40). We showed that these mutations are not part of the previously postulated pathways, and gene-gene interaction analysis found no enrichment of interacting genes in this group (P = 0.57). Also, we were able to show that the de novo mutations in ALS patients are located in genes already prone for de novo mutations (P < 1 × 10-15 ). Although the individual effect of rare de novo mutations in specific genes could not be assessed, our results indicate that, in contrast to previous hypothesis, de novo mutations in general do not impose a major burden on ALS risk.
Keywords: ALS; amyotrophic lateral sclerosis; de novo mutations; disease pathway; motor neuron disease; trios.
© 2017 Wiley Periodicals, Inc.
Conflict of interest statement
DISCLOSURE STATEMENT
The authors declare that they have no conflict of interest.
Figures
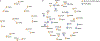
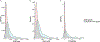
References
-
- Brenner D, Müller K, Wieland T, Weydt P, Böhm S, Lulé D, … Weishaupt JH (2016). NEK1 mutations in familial amyotrophic lateral sclerosis. Brain, 139, e28–e28. - PubMed
-
- Brooks BR, Miller RG, Swash M, Munsat TL, & World Federation of Neurology Research Group on Motor Neuron Diseases. (2000). El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis and Other Motor Neuron Disorders, 1, 293–299. - PubMed
-
- de Jong SW, Huisman MHB, Hennekam EAM, Sutedja NA, van der Kooi AJ, de Visser M, … van den Berg LH (2013). Parental age and the risk of amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration, 14, 224–227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

