Transancestral mapping and genetic load in systemic lupus erythematosus
- PMID: 28714469
- PMCID: PMC5520018
- DOI: 10.1038/ncomms16021
Transancestral mapping and genetic load in systemic lupus erythematosus
Abstract
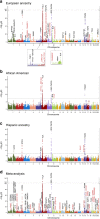
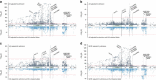
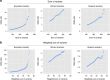
Systemic lupus erythematosus (SLE) is an autoimmune disease with marked gender and ethnic disparities. We report a large transancestral association study of SLE using Immunochip genotype data from 27,574 individuals of European (EA), African (AA) and Hispanic Amerindian (HA) ancestry. We identify 58 distinct non-HLA regions in EA, 9 in AA and 16 in HA (∼50% of these regions have multiple independent associations); these include 24 novel SLE regions (P<5 × 10-8), refined association signals in established regions, extended associations to additional ancestries, and a disentangled complex HLA multigenic effect. The risk allele count (genetic load) exhibits an accelerating pattern of SLE risk, leading us to posit a cumulative hit hypothesis for autoimmune disease. Comparing results across the three ancestries identifies both ancestry-dependent and ancestry-independent contributions to SLE risk. Our results are consistent with the unique and complex histories of the populations sampled, and collectively help clarify the genetic architecture and ethnic disparities in SLE.
Conflict of interest statement
R.R.G., T.B. and T.W.B. are employees of Genentech, Inc. The remaining authors declare no competing financial interests.
Figures


References
-
- Danchenko N., Satia J. A. & Anthony M. S. Epidemiology of systemic lupus erythematosus: a comparison of worldwide disease burden. Lupus 15, 308–318 (2006). - PubMed
-
- Hom G. et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM–ITGAX. N. Engl. J. Med. 358, 900–909 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 GM104938/GM/NIGMS NIH HHS/United States
- R21 AI107176/AI/NIAID NIH HHS/United States
- R01 DK107502/DK/NIDDK NIH HHS/United States
- R01 MD007909/MD/NIMHD NIH HHS/United States
- P30 AR070155/AR/NIAMS NIH HHS/United States
- U54 TR001018/TR/NCATS NIH HHS/United States
- R01 AR063124/AR/NIAMS NIH HHS/United States
- P60 AR062755/AR/NIAMS NIH HHS/United States
- P60 AR053308/AR/NIAMS NIH HHS/United States
- N01 AR022265/AR/NIAMS NIH HHS/United States
- U19 AI082714/AI/NIAID NIH HHS/United States
- P30 GM103510/GM/NIGMS NIH HHS/United States
- R21 AR065626/AR/NIAMS NIH HHS/United States
- P50 AR055503/AR/NIAMS NIH HHS/United States
- P30 AR053483/AR/NIAMS NIH HHS/United States
- R01 AR056360/AR/NIAMS NIH HHS/United States
- K01 AR067280/AR/NIAMS NIH HHS/United States
- RC1 AR058621/AR/NIAMS NIH HHS/United States
- P30 GM110766/GM/NIGMS NIH HHS/United States
- R01 AR043814/AR/NIAMS NIH HHS/United States
- P30 AR072582/AR/NIAMS NIH HHS/United States
- R01 AR057172/AR/NIAMS NIH HHS/United States
- U01 AI101934/AI/NIAID NIH HHS/United States
- R01 AR060366/AR/NIAMS NIH HHS/United States
- P01 AR048929/AR/NIAMS NIH HHS/United States
- UL1 RR029882/RR/NCRR NIH HHS/United States
- R01 AR043727/AR/NIAMS NIH HHS/United States
- R01 CA141700/CA/NCI NIH HHS/United States
- P30 AR070549/AR/NIAMS NIH HHS/United States
- RC2 AR058959/AR/NIAMS NIH HHS/United States
- P01 AI083194/AI/NIAID NIH HHS/United States
- P01 AR049084/AR/NIAMS NIH HHS/United States
- P30 AR047363/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
- Andalusian Health Repository - access to free full text
- Digital CSIC Spanish National Research Council - Access Free Full Text
- Diposit Digital de la Universitat de Barcelona - Access Free Full Text
- Europe PubMed Central
- Linkoping University Electronic Press
- Nature Publishing Group
- PubMed Central
- eScholarship, University of California - Access Free Full Text
Other Literature Sources
Medical
Research Materials

