Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition
- PMID: 28714973
- PMCID: PMC5524181
- DOI: 10.1038/nplants.2017.105
Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition
Erratum in
-
Author Correction: Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition.Nat Plants. 2021 May;7(5):706. doi: 10.1038/s41477-021-00924-y. Nat Plants. 2021. PMID: 33947986
Abstract
The phytohormone auxin induces or represses growth depending on its concentration and the underlying tissue type. However, it remains unknown how auxin signalling is modulated to allow tissues transiting between repression and promotion of growth. Here, we used apical hook development as a model for growth transitions in plants. A PIN-FORMED (PIN)-dependent intercellular auxin transport module defines an auxin maximum that is causal for growth repression during the formation of the apical hook. Our data illustrate that growth transition for apical hook opening is largely independent of this PIN module, but requires the PIN-LIKES (PILS) putative auxin carriers at the endoplasmic reticulum. PILS proteins reduce nuclear auxin signalling in the apical hook, leading to the de-repression of growth and the onset of hook opening. We also show that the phytochrome (phy) B-reliant light-signalling pathway directly regulates PILS gene activity, thereby enabling light perception to repress nuclear auxin signalling and to control growth. We propose a novel mechanism, in which PILS proteins allow external signals to alter tissue sensitivity to auxin, defining differential growth rates.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
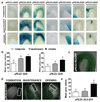
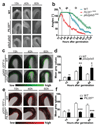
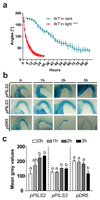
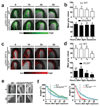

References
-
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. - PubMed
-
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. - PubMed
-
- Rosquete MR, Barbez E, Kleine-Vehn J. Cellular auxin homeostasis: gatekeeping is housekeeping. Molecular plant. 2012;5:772–786. - PubMed
-
- Evans ML, Ishikawa H, Estelle MA. Responses of Arabidopsis roots to auxin studied with high temporal resolution: comparison of wild type and auxin-response mutants. Planta. 1994;194:215–222.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

