Translation is actively regulated during the differentiation of CD8+ effector T cells
- PMID: 28714979
- PMCID: PMC5937989
- DOI: 10.1038/ni.3795
Translation is actively regulated during the differentiation of CD8+ effector T cells
Abstract
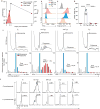
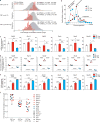
Translation is a critical process in protein synthesis, but translational regulation in antigen-specific T cells in vivo has not been well defined. Here we have characterized the translatome of virus-specific CD8+ effector T cells (Teff cells) during acute infection of mice with lymphocytic choriomeningitis virus (LCMV). Antigen-specific T cells exerted dynamic translational control of gene expression that correlated with cell proliferation and stimulation via the T cell antigen receptor (TCR). The translation of mRNAs that encode translation machinery, including ribosomal proteins, was upregulated during the T cell clonal-expansion phase, followed by inhibition of the translation of those transcripts when the CD8+ Teff cells stopped dividing just before the contraction phase. That translational suppression was more pronounced in terminal effector cells than in memory precursor cells and was regulated by antigenic stimulation and signals from the kinase mTOR. Our studies show that translation of transcripts encoding ribosomal proteins is regulated during the differentiation of CD8+ Teff cells and might have a role in fate 'decisions' involved in the formation of memory cells.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




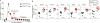
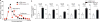
References
-
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27(4):670–684. - PubMed
-
- Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111(6):837–851. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

