Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis
- PMID: 28715499
- PMCID: PMC5531669
- DOI: 10.1371/journal.ppat.1006514
Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis
Abstract
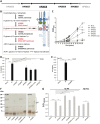
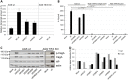
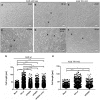
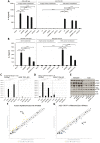
Highly virulent Helicobacter pylori cause proinflammatory signaling inducing the transcriptional activation and secretion of cytokines such as IL-8 in epithelial cells. Responsible in part for this signaling is the cag pathogenicity island (cagPAI) that codetermines the risk for pathological sequelae of an H. pylori infection such as gastric cancer. The Cag type IV secretion system (CagT4SS), encoded on the cagPAI, can translocate various molecules into cells, the effector protein CagA, peptidoglycan metabolites and DNA. Although these transported molecules are known to contribute to cellular responses to some extent, a major part of the cagPAI-induced signaling leading to IL-8 secretion remains unexplained. We report here that biosynthesis of heptose-1,7-bisphosphate (HBP), an important intermediate metabolite of LPS inner heptose core, contributes in a major way to the H. pylori cagPAI-dependent induction of proinflammatory signaling and IL-8 secretion in human epithelial cells. Mutants defective in the genes required for synthesis of HBP exhibited a more than 95% reduction of IL-8 induction and impaired CagT4SS-dependent cellular signaling. The loss of HBP biosynthesis did not abolish the ability to translocate CagA. The human cellular adaptor TIFA, which was described before to mediate HBP-dependent activity in other Gram-negative bacteria, was crucial in the cagPAI- and HBP pathway-induced responses by H. pylori in different cell types. The active metabolite was present in H. pylori lysates but not enriched in bacterial supernatants. These novel results advance our mechanistic understanding of H. pylori cagPAI-dependent signaling mediated by intracellular pattern recognition receptors. They will also allow to better dissect immunomodulatory activities by H. pylori and to improve the possibilities of intervention in cagPAI- and inflammation-driven cancerogenesis.
Conflict of interest statement
The authors have declared no competing interests exist.
Figures
References
-
- Huang JY, Sweeney EG, Sigal M, Zhang HC, Remington SJ, Cantrell MA et al. Chemodetection and Destruction of Host Urea Allows Helicobacter pylori to Locate the Epithelium. Cell Host Microbe 2015; 18(2):147–156. doi: 10.1016/j.chom.2015.07.002 - DOI - PMC - PubMed
-
- Tan S, Tompkins LS, Amieva MR. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog 2009; 5(5):e1000407 doi: 10.1371/journal.ppat.1000407 - DOI - PMC - PubMed
-
- Amieva M, Peek RM Jr., Pathobiology of Helicobacter pylori-Induced Gastric Cancer. Gastroenterology 2016; 150(1):64–78. doi: 10.1053/j.gastro.2015.09.004 - DOI - PMC - PubMed
-
- Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO et al. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci U S A 2004; 101(14):5024–5029. doi: 10.1073/pnas.0308386101 - DOI - PMC - PubMed
-
- Bücker R, Azevedo-Vethacke M, Groll C, Garten D, Josenhans C, Suerbaum S et al. Helicobacter pylori colonization critically depends on postprandial gastric conditions. Sci Rep 2012; 2:994 doi: 10.1038/srep00994 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

