Sulfur K-Edge XAS Studies of the Effect of DNA Binding on the [Fe4S4] Site in EndoIII and MutY
- PMID: 28715891
- PMCID: PMC5568943
- DOI: 10.1021/jacs.7b03966
Sulfur K-Edge XAS Studies of the Effect of DNA Binding on the [Fe4S4] Site in EndoIII and MutY
Abstract
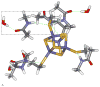
S K-edge X-ray absorption spectroscopy (XAS) was used to study the [Fe4S4] clusters in the DNA repair glycosylases EndoIII and MutY to evaluate the effects of DNA binding and solvation on Fe-S bond covalencies (i.e., the amount of S 3p character mixed into the Fe 3d valence orbitals). Increased covalencies in both iron-thiolate and iron-sulfide bonds would stabilize the oxidized state of the [Fe4S4] clusters. The results are compared to those on previously studied [Fe4S4] model complexes, ferredoxin (Fd), and to new data on high-potential iron-sulfur protein (HiPIP). A limited decrease in covalency is observed upon removal of solvent water from EndoIII and MutY, opposite to the significant increase observed for Fd, where the [Fe4S4] cluster is solvent exposed. Importantly, in EndoIII and MutY, a large increase in covalency is observed upon DNA binding, which is due to the effect of its negative charge on the iron-sulfur bonds. In EndoIII, this change in covalency can be quantified and makes a significant contribution to the observed decrease in reduction potential found experimentally in DNA repair proteins, enabling their HiPIP-like redox behavior.
Figures
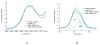
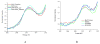






References
-
- Holm RH, Kennepohl P, Solomon EI. Chem Rev. 1996;96:2239–2314. - PubMed
-
- Stiefel EI, George GN. Ferredoxins, Hydrogenases, and Nitrogenases: Metal-Sulfide Proteins. In: Bertini I, editor. Bioinorganic Chemistry. University Science Books; Mill Valley: 1994. pp. 365–453.
-
- Backes G, Mino Y, Loehr TM, Meyer TE, Cusanovich MA, Sweeney WV, Adman ET, Sanders-Loehr J. J Am Chem Soc. 1991;113:2055–2064.
-
- Teo BK, Shulman RG, Brown GS, Meixner AE. J Am Chem Soc. 1979;101:5624.
-
- Czernuszewicz RS, Macor KA, Johnson MK, Gewirth A, Spiro TG. J Am Chem Soc. 1987;109:7178–7187.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

