Biochemical Regulatory Features of Activation-Induced Cytidine Deaminase Remain Conserved from Lampreys to Humans
- PMID: 28716949
- PMCID: PMC5615186
- DOI: 10.1128/MCB.00077-17
Biochemical Regulatory Features of Activation-Induced Cytidine Deaminase Remain Conserved from Lampreys to Humans
Abstract
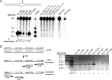
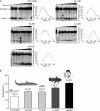
Activation-induced cytidine deaminase (AID) is a genome-mutating enzyme that initiates class switch recombination and somatic hypermutation of antibodies in jawed vertebrates. We previously described the biochemical properties of human AID and found that it is an unusual enzyme in that it exhibits binding affinities for its substrate DNA and catalytic rates several orders of magnitude higher and lower, respectively, than a typical enzyme. Recently, we solved the functional structure of AID and demonstrated that these properties are due to nonspecific DNA binding on its surface, along with a catalytic pocket that predominantly assumes a closed conformation. Here we investigated the biochemical properties of AID from a sea lamprey, nurse shark, tetraodon, and coelacanth: representative species chosen because their lineages diverged at the earliest critical junctures in evolution of adaptive immunity. We found that these earliest-diverged AID orthologs are active cytidine deaminases that exhibit unique substrate specificities and thermosensitivities. Significant amino acid sequence divergence among these AID orthologs is predicted to manifest as notable structural differences. However, despite major differences in sequence specificities, thermosensitivities, and structural features, all orthologs share the unusually high DNA binding affinities and low catalytic rates. This absolute conservation is evidence for biological significance of these unique biochemical properties.
Keywords: biochemistry; enzymes; evolution; evolutionary immunology.
Copyright © 2017 American Society for Microbiology.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
