Genome reconstruction in Cynara cardunculus taxa gains access to chromosome-scale DNA variation
- PMID: 28717205
- PMCID: PMC5514137
- DOI: 10.1038/s41598-017-05085-7
Genome reconstruction in Cynara cardunculus taxa gains access to chromosome-scale DNA variation
Abstract
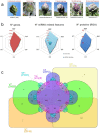
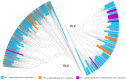
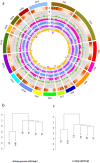
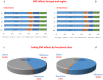
The genome sequence of globe artichoke (Cynara cardunculus L. var. scolymus, 2n = 2x = 34) is now available for use. A survey of C. cardunculus genetic resources is essential for understanding the evolution of the species, carrying out genetic studies and for application of breeding strategies. We report on the resequencing analyses (~35×) of four globe artichoke genotypes, representative of the core varietal types, as well as a genotype of the related taxa cultivated cardoon. The genomes were reconstructed at a chromosomal scale and structurally/functionally annotated. Gene prediction indicated a similar number of genes, while distinctive variations in miRNAs and resistance gene analogues (RGAs) were detected. Overall, 23,5 M SNP/indel were discovered (range 6,34 M -14,50 M). The impact of some missense SNPs on the biological functions of genes involved in the biosynthesis of phenylpropanoid and sesquiterpene lactone secondary metabolites was predicted. The identified variants contribute to infer on globe artichoke domestication of the different varietal types, and represent key tools for dissecting the path from sequence variation to phenotype. The new genomic sequences are fully searchable through independent Jbrowse interfaces (www.artichokegenome.unito.it), which allow the analysis of collinearity and the discovery of genomic variants, thus representing a one-stop resource for C. cardunculus genomics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Wiklund A. The genus Cynara L. (Asteraceae-Cardueae) Bot. J. Linn. Soc. 1992;109:75–123. doi: 10.1111/j.1095-8339.1992.tb00260.x. - DOI
-
- Foury, C. Quelques aspects du développement de l′artichaut (Cynara scolymus L.) issu de semences; analyse plus particulière de la floraison en conditions naturelles. (Paris: VI University Orsay, 1987).
-
- Acquadro A, Portis E, Albertini E, Lanteri S. M-AFLP-based protocol for microsatellite loci isolation in Cynara cardunculus L. (Asteraceae) Mol. Ecol. Notes. 2005;5:272–274. doi: 10.1111/j.1471-8286.2005.00897.x. - DOI
-
- Mauro R, et al. Genetic diversity of globe artichoke landraces from Sicilian small-holdings: implications for evolution and domestication of the species. Conserv. Genet. 2009;10:431–440. doi: 10.1007/s10592-008-9621-2. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

