Isolation of membrane vesicles from prokaryotes: a technical and biological comparison reveals heterogeneity
- PMID: 28717421
- PMCID: PMC5505020
- DOI: 10.1080/20013078.2017.1324731
Isolation of membrane vesicles from prokaryotes: a technical and biological comparison reveals heterogeneity
Abstract
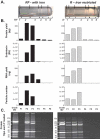
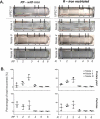
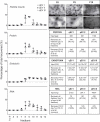
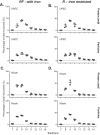
Prokaryotes release membrane vesicles (MVs) with direct roles in disease pathogenesis. MVs are heterogeneous when isolated from bacterial cultures so Density Gradient Centrifugation (DGC) is valuable for separation of MV subgroups from contaminating material. Here we report the technical variability and natural biological heterogeneity seen between DGC preparations of MVs for Mycobacterium smegmatis and Escherichia coli and compare these DGC data with size exclusion chromatography (SEC) columns. Crude preparations of MVs, isolated from cultures by ultrafiltration and ultracentrifugation were separated by DGC with fractions manually collected as guided by visible bands. Yields of protein, RNA and endotoxin, protein banding and particle counts were analysed in these. DGC and SEC methods enabled separation of molecularly distinct MV populations from crude MVs. DGC banding profiles were unique for each of the two species of bacteria tested and further altered by changing culture conditions, for example with iron supplementation. SEC is time efficient, reproducible and cost effective method that may also allow partial LPS removal from Gram-negative bacterial MVs. In summary, both DGC and SEC are suitable for the separation of mixed populations of MVs and we advise trials of both, coupled with complete molecular and single vesicle characterisation prior to downstream experimentation.
Keywords: Extracellular vesicles; microbe; nucleic acids; outer membrane vesicles; pathogen.
Figures

References
-
- Kulkarni HM, Jagannadham MV. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology. 2014;160(Pt 10):2109–2121. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

