Immunogenomic approaches to understand the function of immune disease variants
- PMID: 28718505
- PMCID: PMC5680056
- DOI: 10.1111/imm.12796
Immunogenomic approaches to understand the function of immune disease variants
Abstract
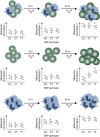
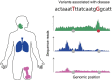
Mapping hundreds of genetic variants through genome wide association studies provided an opportunity to gain insights into the pathobiology of immune-mediated diseases. However, as most of the disease variants fall outside the gene coding sequences the functional interpretation of the exact role of the associated variants remains to be determined. The integration of disease-associated variants with large scale genomic maps of cell-type-specific gene regulation at both chromatin and transcript levels deliver examples of functionally prioritized causal variants and genes. In particular, the enrichment of disease variants with histone marks can point towards the cell types most relevant to disease development. Furthermore, chromatin contact maps that link enhancers to promoter regions in a direct way allow the identification of genes that can be regulated by the disease variants. Candidate genes implicated with such approaches can be further examined through the correlation of gene expression with genotypes. Additionally, in the context of immune-mediated diseases it is important to combine genomics with immunology approaches. Genotype correlations with the immune system as a whole, as well as with cellular responses to different stimuli, provide a valuable platform for understanding the functional impact of disease-associated variants. The intersection of immunogenomic resources with disease-associated variants paints a detailed picture of disease causal mechanisms. Here, we provide an overview of recent studies that combine these approaches to identify disease vulnerable pathways.
Keywords: activation; autoimmunity; cell activation; genomics.
© 2017 The Authors. Immunology Published by John Wiley & Sons Ltd.
Figures

References
-
- Hayter SM, Cook MC. Updated assessment of the prevalence, spectrum and case definition of autoimmune disease. Autoimmun Rev 2012; 11:754–65. - PubMed
-
- Todd JA, Bell JI, McDevitt HO. HLA‐DQ β gene contributes to susceptibility and resistance to insulin‐dependent diabetes mellitus. Nature 1987; 329:599–604. - PubMed
-
- Nepom GT. Major histocompatibility complex‐directed susceptibility to rheumatoid arthritis. Adv Immunol 1998; 68:315–32. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

