Tissue and cell-specific transcriptomes in cotton reveal the subtleties of gene regulation underlying the diversity of plant secondary cell walls
- PMID: 28720072
- PMCID: PMC5516393
- DOI: 10.1186/s12864-017-3902-4
Tissue and cell-specific transcriptomes in cotton reveal the subtleties of gene regulation underlying the diversity of plant secondary cell walls
Erratum in
-
Correction to: Tissue and cell-specific transcriptomes in cotton reveal the subtleties of gene regulation underlying the diversity of plant secondary cell walls.BMC Genomics. 2018 Apr 17;19(1):261. doi: 10.1186/s12864-018-4634-9. BMC Genomics. 2018. PMID: 29665776 Free PMC article.
Abstract
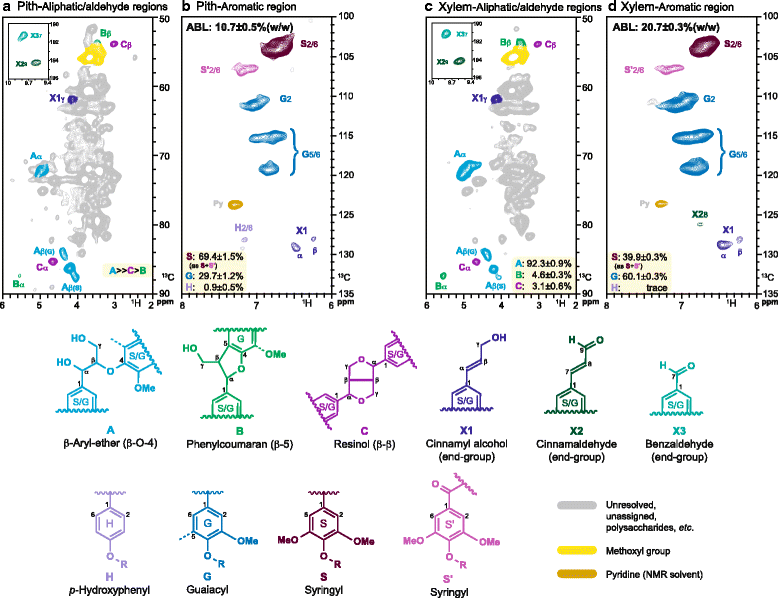
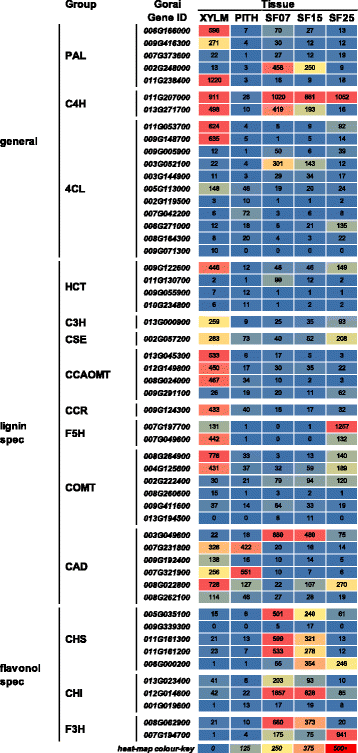
Background: Knowledge of plant secondary cell wall (SCW) regulation and deposition is mainly based on the Arabidopsis model of a 'typical' lignocellulosic SCW. However, SCWs in other plants can vary from this. The SCW of mature cotton seed fibres is highly cellulosic and lacks lignification whereas xylem SCWs are lignocellulosic. We used cotton as a model to study different SCWs and the expression of the genes involved in their formation via RNA deep sequencing and chemical analysis of stem and seed fibre.
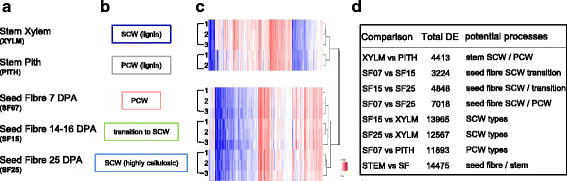
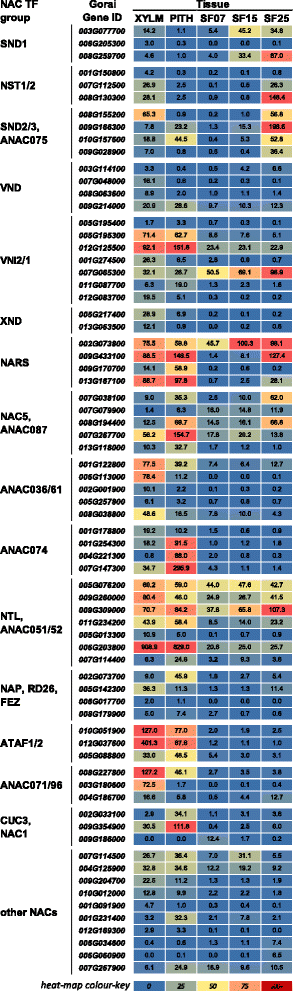
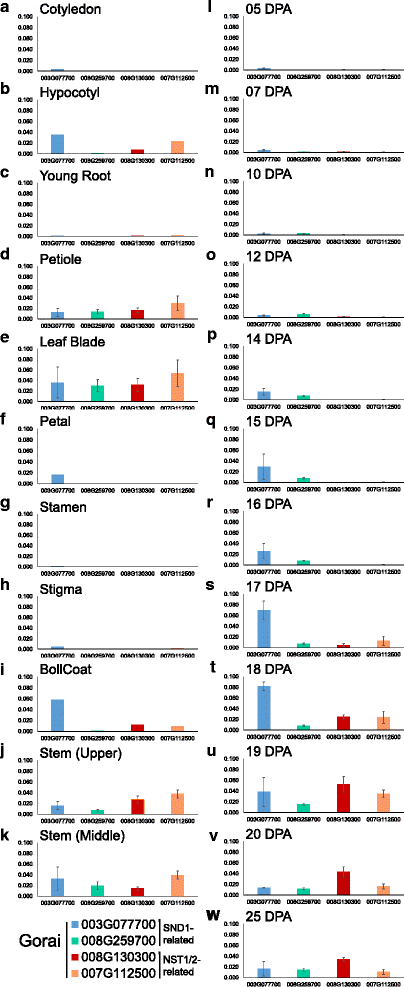
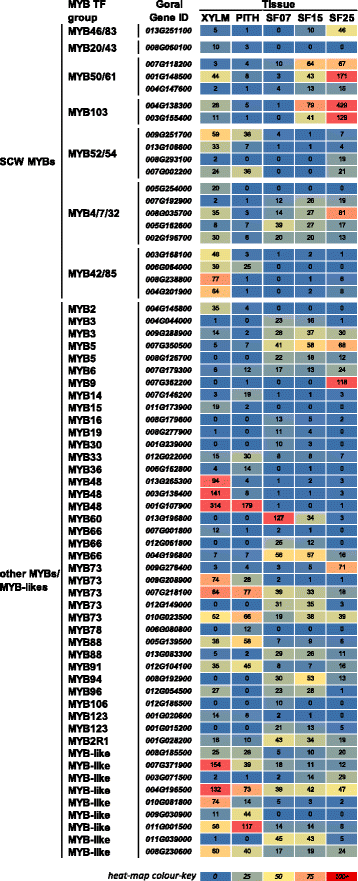
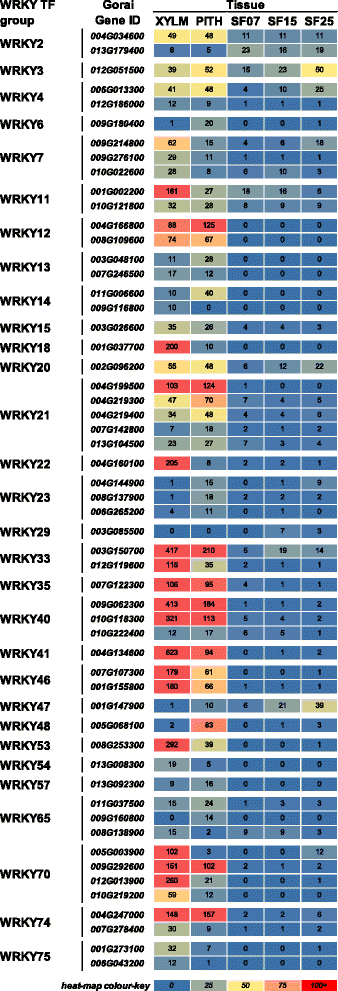
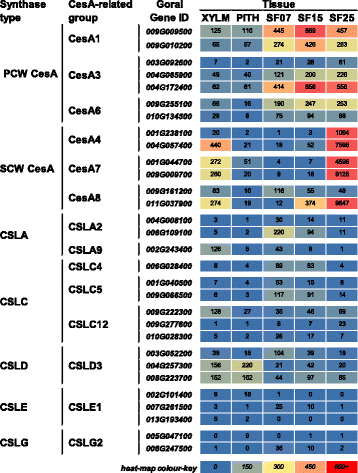
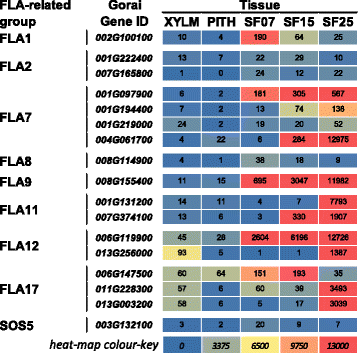
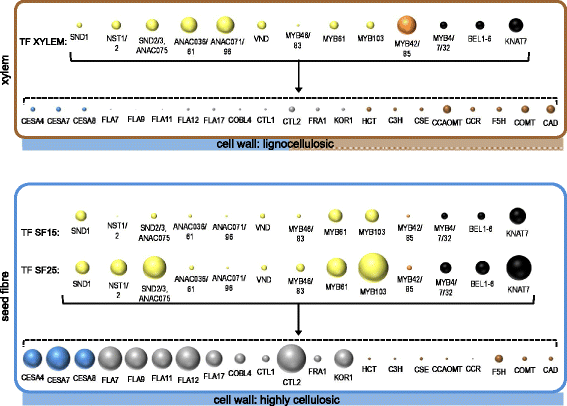
Results: Transcriptome comparisons from cotton xylem and pith as well as from a developmental series of seed fibres revealed tissue-specific and developmentally regulated expression of several NAC transcription factors some of which are likely to be important as top tier regulators of SCW formation in xylem and/or seed fibre. A so far undescribed hierarchy was identified between the top tier NAC transcription factors SND1-like and NST1/2 in cotton. Key SCW MYB transcription factors, homologs of Arabidopsis MYB46/83, were practically absent in cotton stem xylem. Lack of expression of other lignin-specific MYBs in seed fibre relative to xylem could account for the lack of lignin deposition in seed fibre. Expression of a MYB103 homolog correlated with temporal expression of SCW CesAs and cellulose synthesis in seed fibres. FLAs were highly expressed and may be important structural components of seed fibre SCWs. Finally, we made the unexpected observation that cell walls in the pith of cotton stems contained lignin and had a higher S:G ratio than in xylem, despite that tissue's lacking many of the gene transcripts normally associated with lignin biosynthesis.
Conclusions: Our study in cotton confirmed some features of the currently accepted gene regulatory cascade for 'typical' plant SCWs, but also revealed substantial differences, especially with key downstream NACs and MYBs. The lignocellulosic SCW of cotton xylem appears to be achieved differently from that in Arabidopsis. Pith cell walls in cotton stems are compositionally very different from that reported for other plant species, including Arabidopsis. The current definition of a 'typical' primary or secondary cell wall might not be applicable to all cell types in all plant species.
Keywords: Cellulose synthase; Cotton; Gossypium hirsutum; Guaiacyl; Lignin; Primary cell wall; Secondary cell wall; Syringyl; Transcription factor; p-Hydroxyphenyl.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

