Association between genome-wide copy number variation and arsenic-induced skin lesions: a prospective study
- PMID: 28720099
- PMCID: PMC5516382
- DOI: 10.1186/s12940-017-0283-8
Association between genome-wide copy number variation and arsenic-induced skin lesions: a prospective study
Abstract
Background: Exposure to arsenic in drinking water is a global health problem and arsenic-induced skin lesions are hallmark of chronic arsenic toxicity. We and others have reported germline genetic variations as risk factors for such skin lesions. The role of copy number variation (CNV) in the germline DNA in this regard is unknown.
Methods: From a large prospectively followed-up cohort, exposed to arsenic, we randomly selected 2171 subjects without arsenic-induced skin lesions at enrollment and genotyped their whole blood DNA samples on Illumina Cyto12v2.1 SNP chips to generate DNA copy number. Participants were followed up every 2 years for a total of 8 years, especially for the development of skin lesions. In Cox regression models, each CNV segment was used as a predictor, accounting for other potential covariates, for incidence of skin lesions.
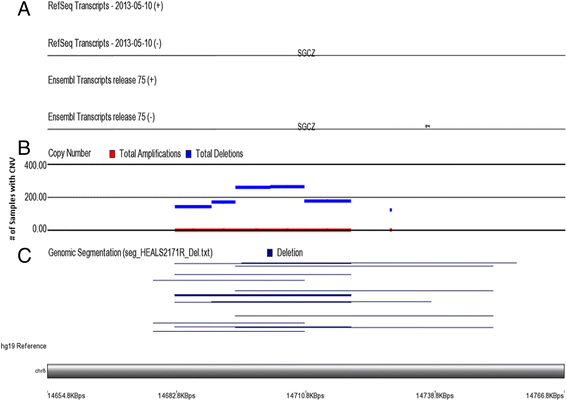
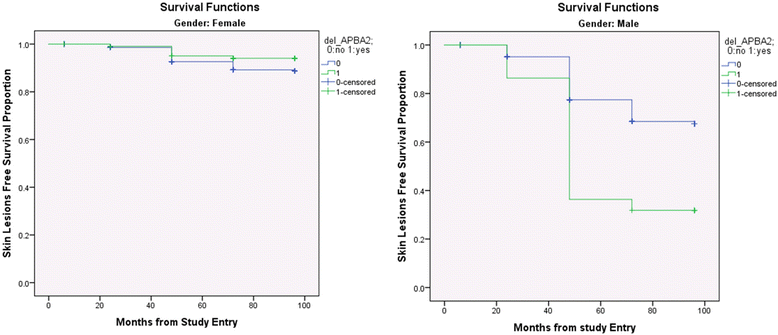
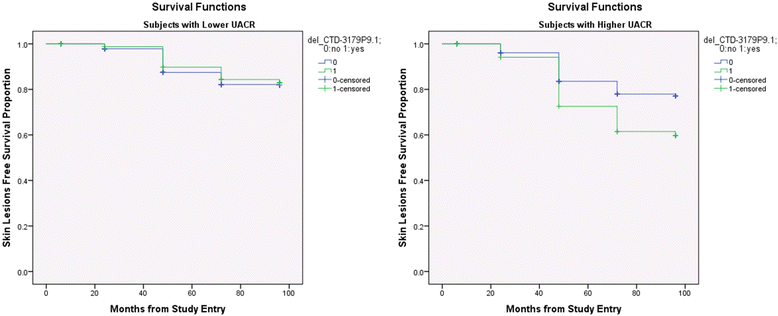
Result: The presence of genomic deletion(s) in a number of genes (OR5J2, GOLGA6L7P, APBA2, GALNTL5, VN1R31P, PHKG1P2, SGCZ, ZNF658) and lincRNA genes (RP11-76I14.1, CTC-535 M15.2, RP11-73B2.2) were associated with higher risk [HR between 1.67 (CI 1.3-2.1) and 2.15 (CI 1.5-2.9) for different CNVs] for development of skin lesions independent of gender, age, and arsenic exposure. Some deletions had stronger effect in a specific gender (ZNF658 in males, SGCZ in females) and some had stronger effect in higher arsenic exposure (lincRNA CTD-3179P9.1) suggesting a possible gene-environment interaction.
Conclusion: This first genome-wide CNV study in a prospectively followed-up large cohort, exposed to arsenic, suggests that DNA deletion in several genes and lincRNA genes may predispose an individual to a higher risk of development of arsenic-induced skin lesions.
Keywords: Arsenic; Copy number variation; Gene-environment interaction; Skin lesion; Survival analysis; lincRNA.
Conflict of interest statement
Ethics approval and consent to participate
The study protocol was approved by the Institutional Review board of The University of Chicago, Columbia University, and the Bangladesh Medical Research Council. Informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no actual or potential competing financial interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- United States Environmental Protection Agency: Office of Water (4606) drinking water standard for arsenic. EPA 815-F-00-015. In.; 2001.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

