PacBio assembly of a Plasmodium knowlesi genome sequence with Hi-C correction and manual annotation of the SICAvar gene family
- PMID: 28720171
- PMCID: PMC5798397
- DOI: 10.1017/S0031182017001329
PacBio assembly of a Plasmodium knowlesi genome sequence with Hi-C correction and manual annotation of the SICAvar gene family
Abstract
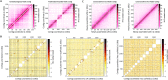
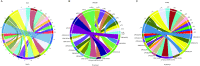
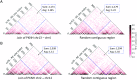
Plasmodium knowlesi has risen in importance as a zoonotic parasite that has been causing regular episodes of malaria throughout South East Asia. The P. knowlesi genome sequence generated in 2008 highlighted and confirmed many similarities and differences in Plasmodium species, including a global view of several multigene families, such as the large SICAvar multigene family encoding the variant antigens known as the schizont-infected cell agglutination proteins. However, repetitive DNA sequences are the bane of any genome project, and this and other Plasmodium genome projects have not been immune to the gaps, rearrangements and other pitfalls created by these genomic features. Today, long-read PacBio and chromatin conformation technologies are overcoming such obstacles. Here, based on the use of these technologies, we present a highly refined de novo P. knowlesi genome sequence of the Pk1(A+) clone. This sequence and annotation, referred to as the 'MaHPIC Pk genome sequence', includes manual annotation of the SICAvar gene family with 136 full-length members categorized as type I or II. This sequence provides a framework that will permit a better understanding of the SICAvar repertoire, selective pressures acting on this gene family and mechanisms of antigenic variation in this species and other pathogens.
Keywords: Plasmodium knowlesi; SICAvar; Hi-C; MaHPIC; PacBio; annotation; antigenic variation; genome; sequence.
Figures
Similar articles
-
A SICAvar switching event in Plasmodium knowlesi is associated with the DNA rearrangement of conserved 3' non-coding sequences.Mol Biochem Parasitol. 2004 Nov;138(1):37-49. doi: 10.1016/j.molbiopara.2004.05.017. Mol Biochem Parasitol. 2004. PMID: 15500914
-
Spleen-dependent regulation of antigenic variation in malaria parasites: Plasmodium knowlesi SICAvar expression profiles in splenic and asplenic hosts.PLoS One. 2013 Oct 18;8(10):e78014. doi: 10.1371/journal.pone.0078014. eCollection 2013. PLoS One. 2013. PMID: 24205067 Free PMC article.
-
Antigenic variation in malaria: a 3' genomic alteration associated with the expression of a P. knowlesi variant antigen.Mol Cell. 1999 Feb;3(2):131-41. doi: 10.1016/s1097-2765(00)80304-4. Mol Cell. 1999. PMID: 10078196
-
Plasmodium knowlesi: a superb in vivo nonhuman primate model of antigenic variation in malaria.Parasitology. 2018 Jan;145(1):85-100. doi: 10.1017/S0031182017001135. Epub 2017 Jul 17. Parasitology. 2018. PMID: 28712361 Free PMC article. Review.
-
Variant antigen expression in malaria infections: posttranscriptional gene silencing, virulence and severe pathology.Mol Biochem Parasitol. 2004 Mar;134(1):17-25. doi: 10.1016/j.molbiopara.2003.09.013. Mol Biochem Parasitol. 2004. PMID: 14747139 Review. No abstract available.
Cited by
-
Population genomics in neglected malaria parasites.Front Microbiol. 2022 Sep 8;13:984394. doi: 10.3389/fmicb.2022.984394. eCollection 2022. Front Microbiol. 2022. PMID: 36160257 Free PMC article. Review.
-
Plasmodium knowlesi Cytoadhesion Involves SICA Variant Proteins.Front Cell Infect Microbiol. 2022 Jun 23;12:888496. doi: 10.3389/fcimb.2022.888496. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35811680 Free PMC article.
-
Decrypting the complexity of the human malaria parasite biology through systems biology approaches.Front Syst Biol. 2022;2:940321. doi: 10.3389/fsysb.2022.940321. Epub 2022 Sep 16. Front Syst Biol. 2022. PMID: 37200864 Free PMC article.
-
Clinical recovery of Macaca fascicularis infected with Plasmodium knowlesi.Malar J. 2021 Dec 30;20(1):486. doi: 10.1186/s12936-021-03925-6. Malar J. 2021. PMID: 34969401 Free PMC article.
-
The Genome of Plasmodium gonderi: Insights into the Evolution of Human Malaria Parasites.Genome Biol Evol. 2024 Feb 1;16(2):evae027. doi: 10.1093/gbe/evae027. Genome Biol Evol. 2024. PMID: 38376987 Free PMC article.
References
-
- al-Khedery B., Barnwell J. W. and Galinski M. R. (1999). Antigenic variation in malaria: a 3′ genomic alteration associated with the expression of a P. knowlesi variant antigen. Molecular Cell 3, 131–141. - PubMed
-
- Assefa S., Lim C., Preston M. D., Duffy C. W., Nair M. B., Adroub S. A., Kadir K. A., Goldberg J. M., Neafsey D. E., Divis P., Clark T. G., Duraisingh M. T., Conway D. J., Pain A. and Singh B. (2015). Population genomic structure and adaptation in the zoonotic malaria parasite Plasmodium knowlesi. Proceedings of the National Academy of Sciences of the USA 112, 13027–13032. - PMC - PubMed
-
- Aurrecoechea C., Barreto A., Basenko E. Y., Brestelli J., Brunk B. P., Cade S., Crouch K., Doherty R., Falke D., Fischer S., Gajria B., Harb O. S., Heiges M., Hertz-Fowler C., Hu S., Iodice J., Kissinger J. C., Lawrence C., Li W., Pinney D. F., Pulman J. A., Roos D. S., Shanmugasundram A., Silva-Franco F., Steinbiss S., Stoeckert C. J. Jr., Spruill D., Wang H., Warrenfeltz S. and Zheng J. (2017). EuPathDB: the eukaryotic pathogen genomics database resource. Nucleic Acids Research 45(D1), D581–D591. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

