Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131
- PMID: 28720578
- PMCID: PMC5538559
- DOI: 10.1101/gr.216606.116
Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131
Abstract
Escherichia coli associated with urinary tract infections and bacteremia has been intensively investigated, including recent work focusing on the virulent, globally disseminated, multidrug-resistant lineage ST131. To contextualize ST131 within the broader E. coli population associated with disease, we used genomics to analyze a systematic 11-yr hospital-based survey of E. coli associated with bacteremia using isolates collected from across England by the British Society for Antimicrobial Chemotherapy and from the Cambridge University Hospitals NHS Foundation Trust. Population dynamics analysis of the most successful lineages identified the emergence of ST131 and ST69 and their establishment as two of the five most common lineages along with ST73, ST95, and ST12. The most frequently identified lineage was ST73. Compared to ST131, ST73 was susceptible to most antibiotics, indicating that multidrug resistance was not the dominant reason for prevalence of E. coli lineages in this population. Temporal phylogenetic analysis of the emergence of ST69 and ST131 identified differences in the dynamics of emergence and showed that expansion of ST131 in this population was not driven by sequential emergence of increasingly resistant subclades. We showed that over time, the E. coli population was only transiently disturbed by the introduction of new lineages before a new equilibrium was rapidly achieved. Together, these findings suggest that the frequency of E. coli lineages in invasive disease is driven by negative frequency-dependent selection occurring outside of the hospital, most probably in the commensal niche, and that drug resistance is not a primary determinant of success in this niche.
© 2017 Kallonen et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
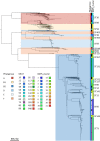
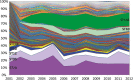



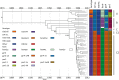

Comment in
-
Pathogen genetics: Evolutionary dynamics driving drug resistance.Nat Rev Genet. 2017 Oct;18(10):578-579. doi: 10.1038/nrg.2017.68. Epub 2017 Aug 14. Nat Rev Genet. 2017. PMID: 28804137 No abstract available.
References
-
- Alhashash F, Wang X, Paszkiewicz K, Diggle M, Zong Z, McNally A. 2016. Increase in bacteraemia cases in the East Midlands region of the UK due to MDR Escherichia coli ST73: high levels of genomic and plasmid diversity in causative isolates. J Antimicrob Chemother 71: 339–343. - PubMed
-
- Allsopp LP, Totsika M, Tree JJ, Ulett GC, Mabbett AN, Wells TJ, Kobe B, Beatson SA, Schembri MA. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect Immun 78: 1659–1669. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
