EWS/FLI is a Master Regulator of Metabolic Reprogramming in Ewing Sarcoma
- PMID: 28720588
- PMCID: PMC5668171
- DOI: 10.1158/1541-7786.MCR-17-0182
EWS/FLI is a Master Regulator of Metabolic Reprogramming in Ewing Sarcoma
Abstract
Ewing sarcoma is a bone malignancy driven by a translocation event resulting in the fusion protein EWS/FLI1 (EF). EF functions as an aberrant and oncogenic transcription factor that misregulates the expression of thousands of genes. Previous work has focused principally on determining important transcriptional targets of EF, as well as characterizing important regulatory partnerships in EF-dependent transcriptional programs. Less is known, however, about EF-dependent metabolic changes or their role in Ewing sarcoma biology. Therefore, the metabolic effects of silencing EF in Ewing sarcoma cells were determined. Metabolomic analyses revealed distinct separation of metabolic profiles in EF-knockdown versus control-knockdown cells. Mitochondrial stress tests demonstrated that knockdown of EF increased respiratory as well as glycolytic functions. Enzymes and metabolites in several metabolic pathways were altered, including de novo serine synthesis and elements of one-carbon metabolism. Furthermore, phosphoglycerate dehydrogenase (PHGDH) was found to be highly expressed in Ewing sarcoma and correlated with worse patient survival. PHGDH knockdown or pharmacologic inhibition in vitro caused impaired proliferation and cell death. Interestingly, PHGDH modulation also led to elevated histone expression and methylation. These studies demonstrate that the translocation-derived fusion protein EF is a master regulator of metabolic reprogramming in Ewing sarcoma, diverting metabolites toward biosynthesis. As such, these data suggest that the metabolic aberrations induced by EF are important contributors to the oncogenic biology of these tumors.Implications: This previously unexplored role of EWS/FLI1-driven metabolic changes expands the understanding of Ewing sarcoma biology, and has potential to significantly inform development of therapeutic strategies. Mol Cancer Res; 15(11); 1517-30. ©2017 AACR.
©2017 American Association for Cancer Research.
Figures

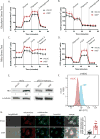
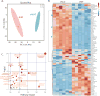
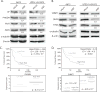

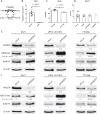
References
-
- Lahl M, Fisher VL, Laschinger K. Ewing’s sarcoma family of tumors: an overview from diagnosis to survivorship. Clin J Oncol Nurs. 2008;12:89–97. - PubMed
-
- Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle Georget Tex. 2008;7:250–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

